Acknowledgements
The authors would like to thank the staff who were involved in the successful delivery of this project from UK Health Security Agency, Animal and Plant Health Agency (APHA), Agri-Food and Biosciences Institute (AFBI) and the Quadram Institute Bioscience through the testing of samples and isolates. We are also grateful to Hallmark Meat Hygiene Ltd. for their collaboration in collecting the salmon samples.
AMR Terminology
Internationally, epidemiological cut-off values (ECOFFs) have been used to determine ‘resistance’ (i.e. susceptibility to antimicrobials), but it should be noted that ECOFFs do not necessarily indicate clinical resistance (i.e. whether a treatment is likely to be unsuccessful). ECOFFs distinguish between individuals within a species which have or have not developed any phenotypically detectable acquired resistance. The ECOFF is the upper end of the wild type[1] of the species for the agent and does not necessarily indicate clinical resistance[2]. The Advisory Committee on Microbiological Safety of Food Working Group on Antimicrobial Resistance (ACMSF AMR WG) has published recommendations on the use of AMR terminology/nomenclature used in FSA reports which includes how best to integrate ECOFFs, clinical breakpoint and genomic data to most accurately reflect the AMR status of bacterial isolates found in food surveys. Results in this study where we have assigned ‘resistance’ are based on ECOFFs.
1. Lay Summary
In 2022, the Veterinary Medicines Directorate (VMD) reported that the usage of antibiotics in the UK farmed salmon industry had increased by 168% between 2017 and 2021. Additionally, there was a Listeria monocytogenes outbreak in 2022 linked to smoked salmon. This raised questions about whether salmon sold in the UK is contaminated with L. monocytogenes and antimicrobial resistant (AMR) bacteria and the risk this poses to consumers. Whilst raw salmon is not considered to be a ready-to-eat (RTE) product, and therefore not required to meet regulatory criteria in relation to L. monocytogenes presence, it may be used in the production of RTE products with no, or limited, heat processes involved, resulting in a risk of L. monocytogenes (and other bacteria) surviving in the finished product. This survey was carried out to generate new data on the presence of L. monocytogenes, E. coli and the levels of AMR found in these organisms in raw, chilled, prepacked farmed salmon fillets on retail sale in the UK.
A total of 307 salmon fillets were collected from major supermarkets within the UK between January and December 2024. These salmon samples were tested for the presence of L. monocytogenes (a foodborne disease-causing bacterium), E. coli (a type of bacteria that can indicate poor hygiene practices) and resistance to a range of antimicrobials in both bacterial species. This survey found that:
-
L. monocytogenes was detected in 1.6% (5 out of 307) of the salmon fillets tested at low levels (less than 20 cfu/g) and was not resistant to common antibiotics.
-
E. coli was detected in 35% (108 out of 307) of the salmon tested, albeit at low levels (107 samples had E. coli levels less than 20 cfu/g with one sample at 20 cfu/g). Of 102 E. coli isolates examined, 4 (4%) were resistant to one type of antibiotic, ampicillin.
-
Importantly, no salmon samples had E. coli or L. monocytogenes that were resistant to key clinically important antibiotics.
-
This study provides good news, as it shows that the likelihood of consumers acquiring L. monocytogenes and AMR from UK retail raw salmon fillets is low. Furthermore, following good hygienic handling and cooking practices will help to minimise the risk even further.
2. Executive Summary
Antimicrobial resistance (AMR) is a significant public health concern, and addressing this threat is a national strategic priority for the UK. The Government’s AMR national action plan for 2024-2029 highlights the importance of improving food safety to limit the contamination and spread of AMR via the food chain and identifies the need to strengthen the evidence base for AMR and food safety through research and surveys, as well as promoting good hygiene practices across the food chain.
In 2022, the Veterinary Medicines Directorate (VMD) published its annual report on veterinary antibiotic sales and usage, which highlighted that, in contrast to many other food-producing animal sectors, antibiotic usage in the salmon industry had increased by 168% between 2017 and 2021. Following the publication of this report, the Food Standard Agency’s Antimicrobial Resistance (AMR) Research and Evidence Programme Review held in March 2023 highlighted the need for more data on antimicrobial resistance in bacteria from fish. Additionally, recent outbreaks of listeriosis associated with smoked fish consumption have raised public health concerns. While antimicrobial resistance is not widespread in Listeria strains in the UK and infections are usually treatable with penicillin or ampicillin, it is beneficial to monitor AMR in this pathogen in order to detect changes in resistance over time.
This study assessed the prevalence of Listeria species and Escherichia coli (E. coli) in raw salmon fillets on retail sale in the UK and analysed the antimicrobial resistance profiles of any L. monocytogenes and E. coli that were detected, thus allowing an assessment of the potential public health risks posed by raw salmon to the consumer.
Between January and December 2024, a total of 307 samples of raw, fresh, prepacked, farmed salmon fillets were collected by HallMark Meat Hygiene Ltd from major supermarkets in all regions of the UK, based on market share data. Samples were tested for the presence of L. monocytogenes, other Listeria species and E. coli including presumptive extended spectrum beta-lactamase (ESBL- and AmpC-) producing-, colistin-resistant and carbapenemase-producing E. coli.
E. coli isolates were tested to determine the Minimum Inhibitory Concentrations (MIC) for a range of antimicrobials. L. monocytogenes isolates were subjected to whole genome sequencing (WGS) and typed by single nucleotide polymorphisms (SNPs) analysis to assess any matching clinical isolates from cases of listeriosis and characterised to identify any AMR determinants.
L. monocytogenes was detected in five samples (1.6%), with these isolates being of sequence types ST7 (clonal complex CC7), ST8 (CC8), ST18 (CC18), ST121 (CC121) and ST425 (CC90) respectively. Other Listeria species were detected in 31 samples (10%), including L. welshimeri in 18 (5.9%) samples, L. innocua in 12 (3.9%) and both L. welshimeri and L. innocua in one sample (0.3%). Four of the L. monocytogenes isolates were from the same retailer but sampled in different regions of the country and at different times (with different batch numbers and different processing establishment codes). Two L. monocytogenes isolates showed genetic similarities with strains linked to human listeriosis cases: the ST121 isolate closely matched an isolate from a human case from August 2023 and the ST8 isolate was similar to strains from four human cases in 2020, based on WGS SNPs analysis. These genetic similarities suggest a common source for the salmon and human strains.
Generic E. coli were detected in 108 samples (35%), but only one of these was at a countable level of 20 cfu/g. Four generic E. coli isolates (3.9%) were resistant to ampicillin, but the remainder were sensitive to all antimicrobials tested. No ESBL/Amp C-producing E. coli, colistin resistant E. coli or carbapenemase-producing E. coli were detected in any salmon samples.
As only a small number of L. monocytogenes were recovered from raw salmon in this study, additional data on AMR resistance was obtained by reviewing WGS results from a further 15 L. monocytogenes strains previously detected in raw salmon samples by the UK Health Security Agency laboratories between 2015 and 2024. All the 20 L. monocytogenes isolates examined (five from this study and 15 others previously tested) contained genes conferring intrinsic resistance to fosfomycin (an antibiotic used in the treatment of osteomyelitis and bacterial meningitis). No other antimicrobial resistance genes were detected. However, all 20 L. monocytogenes isolates carried genes that potentially code for resistance to benzalkonium chloride, a quaternary ammonium compound widely used as a disinfectant in the food industry.
This study of raw salmon fillets collected from supermarkets in the UK confirmed a low prevalence of L. monocytogenes in this commodity, as well as low levels of both generic E. coli and specific antimicrobial-resistant E. coli. In L. monocytogenes isolates both from this study and from a wider range of raw salmon samples examined between 2015 and 2024, AMR was limited to intrinsic fosfomycin resistance only. These results provide reassurance for consumers and regulators of the generally low microbiological (and specifically AMR) risk associated with raw salmon on the market.
3. Introduction
Antimicrobial resistance is a significant public health concern, with an estimated 4.95 million deaths being associated with antimicrobial resistant (AMR) bacteria globally in 2019 (Antimicrobial Resistance Collaborators, 2022). AMR E. coli was responsible for more deaths than any other single pathogen, and in particular, E. coli with resistance to the third-generation cephalosporin antimicrobials or fluoroquinolones were both amongst the top seven pathogen-drug combinations in terms of the numbers of deaths caused (Antimicrobial Resistance Collaborators, 2022).
Increasingly, a ‘One Health’ approach is being taken in relation to understanding and controlling AMR infections, which ensures that the impact of these organisms in humans, animals and the environment are all taken into consideration. Therefore, reducing exposure to antimicrobials unrelated to treating human disease, such as in the production of food animals, is seen as an important aspect of controlling AMR in human infections. This is particularly important given that much of the food consumed in the UK has been imported from other parts of the world, where the use of antimicrobials in agriculture may not be as tightly controlled as it is in the UK and Europe.
The March 2023 review of the Food Standard Agency’s Antimicrobial Resistance (AMR) Research and Evidence Programme highlighted the need for more data on antimicrobial resistance in bacteria from fish (Nicholls et al., 2023). The FSA decided that a survey focussing on AMR in farmed salmon would be beneficial due to evidence that the usage of antimicrobials in UK salmon farming had increased by 168% between 2017 and 2021 (UK-VARSS 2021, 2022) and concerns on whether farmed salmon on sale in the UK was contaminated with AMR and the potential risk this poses to consumers as salmon can be consumed raw or lightly cooked. Farmed salmon is the most popular fish within the UK with Fish Farming Expert estimating that the sales of salmon in 2023 accounted for over 28% of all fish bought in the UK (£4.36 billion) and 14% of 387,463 tonnes of all fish sales over the same period. In order to manage the cost and complexity of the survey, the FSA decided against including other farmed fish species in the study design.
E. coli is frequently used as a marker for the development of AMR in food, including extended spectrum beta-lactamase (ESBL)-producing and carbapenemase-producing bacteria. Historical data is available for AMR in E. coli isolated from a range of food types over many years (APHA, 2021, 2022; Randall et al., 2017, 2020; Willis et al., 2023), and therefore this organism allows comparison of AMR trends over time and between food commodities (and food animals).
Listeria monocytogenes in salmon has become a public health concern due to its association with recent outbreaks of listeriosis linked with smoked salmon consumption. An outbreak of 20 cases of L. monocytogenes ST1607 was reported in Denmark, Germany and Italy between 2019 and 2024, including 5 deaths. Investigations identified the outbreak strain in smoked salmon products produced in Denmark (ECDC-EFSA, 2024b). A further outbreak of 73 cases of L. monocytogenes ST173 in the UK and European countries between 2012 and 2024 was linked to the consumption of fish products (ECDC-EFSA, 2024a). In the UK, 24 cases of listeriosis (clonal complex CC217) were reported between 2020 and 2024 linked to contaminated smoked salmon (FSA, 2024), with four fatalities.
A review of UK listeriosis cases between 2014 and 2023 identified smoked fish as the second most common category associated with L. monocytogenes infection after ready-to-eat sandwiches/salads (UKHSA, 2023) and the FSA and NHS published updated guidance in 2022 advising vulnerable groups of the increased risk of Listeria infection linked to ready-to-eat smoked fish (FSA, 2022; NHS, 2022).
Data available on the prevalence of AMR in L. monocytogenes in the UK is limited, probably due to the relatively low number of reported cases (approximately 160 cases of listeriosis per year in England and Wales between 2013 and 2022, according to UKHSA data; and approximately 14 cases per year in Scotland [Public Health Scotland, 2023]) but also because resistance to treatment is rarely observed.
In a study of 102 raw marine fish in Poland, L. monocytogenes was detected in 18% of samples (Wieczorek & Osek, 2017) with phenotypic resistance to oxacillin (58%), ceftriaxone (32%) and clindamycin (9%) being detected in these isolates. A further study in a Polish fish processing plant detected L. monocytogenes in 32 of 119 raw material samples (27%) (Skowron et al., 2018), and four of these were resistant to at least three antibiotics using a disc diffusion method, including penicillin, ampicillin, meropenem, erythromycin and trimethoprim/ sulfamethoxazole. A Spanish study of 50 isolates of Listeria species from ready-to-eat foods of animal origin found that 90% showed phenotypic resistance to clindamycin, 30% to tetracycline and 26% to ciprofloxacin (Escolar et al., 2017).
This study aimed to provide data on the prevalence of Listeria species and E. coli in raw, prepacked, farmed salmon fillets on retail sale in the UK, and on the AMR profiles of any L. monocytogenes and E. coli that were detected, in order to determine potential public health risks posed by raw salmon to the consumer.
4. Materials and Methods
4.1. Sample collection at retail and transportation to the testing laboratory
Sampling design and collection were completed by HallMark Meat Hygiene Ltd, trading as HallMark Veterinary & Compliance Services. The sampling plan aimed for 315 samples, including a 5% contingency above the recommended 300 samples to account for potential missing or rejected samples. The sampling was conducted over 12 months from January 2024 to December 2024, ensuring seasonal distribution. Samples were evenly allocated across four quarters, with approximately 75 samples per quarter and an estimated 25 samples per month to mitigate seasonal variation and avoid introducing systematic bias associated with time-limited sampling.
HallMark sampling officers were instructed to collect pre-packaged, labelled, fresh (chilled) farmed salmon fillets, covering products from both UK and overseas sources. Frozen, wild, loose, processed, or prepared salmon products (e.g., smoked, marinated, or canned) were excluded.
Sampling was conducted in retail outlets proportionally selected based on market share data provided by the FSA. The selection of sampling regions was based on UK International Territorial Levels (ITL3) regions, ensuring geographic representation. Regions contributing less than 20% of the national population were excluded, with substitutions made where necessary. Retailers were selected based on market share proportions, ensuring proportional representation of major supermarket chains. Samples were collected from the top nine UK retailers, with minor variations from the original sampling plan that did not significantly affect market share representation.
Sampling officers followed consumer-like purchasing behaviours, selecting eligible products within retail outlets while avoiding batch duplication to ensure a broad representation of products.
This study design ensured that the sampling process was statistically robust, geographically representative, and aligned with FSA surveillance objectives.
After collection, samples were packed into cold boxes with sufficient ice packs to achieve and maintain a temperature of <8°C during transit. Samples were dispatched by overnight courier to one of four designated microbiology laboratories: UKHSA Porton FW&E Laboratory, UKHSA London FW&E Laboratory, UKHSA York FW&E Laboratory, or Agri-Food and Biosciences Institute (AFBI), Belfast.
In total, 323 samples were collected, of which 307 samples were classified as satisfactory at the time of receipt at the laboratories, while 16 samples were rejected due to extended transit time, incorrect sample type or duplicated batch number. Given the 5% contingency allowance, the total number of satisfactorily tested samples met the project objectives.
Samples were either tested immediately or stored at 2-8°C and tested within 36 hours (h) of receipt, and within the shelf-life of the product. Once portions had been removed for testing, the remainder of each sample was kept in its original packaging and stored at -18°C.
4.2. Examination of salmon samples for the presence of generic and antimicrobial resistant Escherichia coli
A 10-1 homogenate of each raw salmon sample was prepared by diluting a 27 g aliquot of salmon in Buffered Peptone Water (BPW; prepared in-house), according to ISO 6887-1:2017 (International Organization for Standardization, 2017c). A portion of this homogenate (20 ml) was retained and used to enumerate generic E. coli using surface inoculation of a tryptone bile X-glucuronide (TBX) agar plate (Table 1).
The remaining 250 ml of homogenate was incubated at 37°C for 18 h and then sub-cultured for the detection of generic E. coli and presumptive ESBL-/AmpC- producing E. coli, colistin-resistant E. coli and carbapenemase-producing E. coli (Table 1).
4.3. Examination of salmon samples for the presence of Listeria species
A 25 g portion of salmon was examined for the presence of L. monocytogenes and other Listeria species using an in-house method based on ISO 11290-1:2017 (International Organization for Standardization, 2017a). Briefly, sufficient Half Fraser broth (prepared in-house) was added to prepare a 10-1 dilution. The diluted sample was homogenised in a stomacher and incubated at 30°C for 24 h. The incubated broth was then sub-cultured to Fraser Broth as well as onto Oxford agar (E&O Laboratories) and chromogenic Listeria agar plates (Colorex; E&O Laboratories). Following a further 48 h incubation, the Fraser broth was sub-cultured onto fresh Oxford and OCLA agar plates.
Each sample was also examined for the enumeration of Listeria species by surface inoculation of a chromogenic Listeria agar plate using 0.5 ml of the BPW homogenate produced as described in section 6.2 and incubation for 48 h at 37°C (International Organization for Standardization, 2017b).
All agar plates were incubated for 48 h at 37°C and examined after 24 and 48 h. Suspect colonies were confirmed using a MALDI-TOF instrument (Bruker), and representative L. monocytogenes isolates from each positive sample were retained on nutrient agar slopes and sent to the UKHSA Gastrointestinal Bacteria Reference Unit (GBRU) and Quadram Institute Bioscience for Whole Genome Sequencing.
4.4. Determination of minimum inhibitory concentrations for E. coli isolates
One isolate from each isolation medium for each sample was selected to determine the minimum inhibitory concentrations (MIC) for a range of antimicrobials. A broth microdilution method was used to determine the MICs, using SensititreTM. Isolates were inoculated into Mueller Hinton broth at a suitable concentration and dispensed onto commercially prepared plates (Sensititre EUVSEC3, Thermofisher) containing two-fold dilution series of antimicrobials in accordance with Assimilated Decision 2020/1729/EU. After incubation at 37°C for 18-24 h the plates were examined and growth end-points established for each antimicrobial to provide MICs.
Microbiologically-resistant and susceptible interpretations for the MICs were obtained by comparison with epidemiological cut-off values (ECOFFs) specified in EU Decision 2020/1729 (European Commission, 2020), and if not available then current ECOFFs published by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) were considered (Table 2). ECOFF values separate the naive, susceptible bacterial populations from isolates that have developed reduced susceptibility to a given antimicrobial agent as recommended in the ECDC EU protocol for harmonised monitoring of antimicrobial resistance in human Salmonella spp. and Campylobacter spp. isolates (ECDC, 2016). The ECOFFs differ from breakpoints used for clinical purposes, which are defined against a background of clinically relevant data.
4.5. Characterisation of Listeria monocytogenes isolates using whole genome sequencing and identification of links to human cases
L. monocytogenes isolates were sent to GBRU for further characterisation and typing by WGS as described previously (McLauchlin et al., 2021). Briefly, DNA was obtained by automated extraction (QIAsymphony DSP DNA Kit, Qiagen, Manchester, England) according to manufacturer instructions and sequenced by the UKHSA Central Sequencing Laboratory: sample preparation was using the NexteraXT (Illumina Inc, San Diego, USA) and sequencing was using Illumina HiSeq 2500 platform with 2 × 100 bp reads (Illumina Inc). Short reads were quality trimmed using Trimmomatic (Bolger et al., 2014) removing the sequence adaptor. Clonal complexes (CCs) were derived from WGS analysis and were assigned using MOST (Tewolde et al., 2016) in accordance with the designation of the [Institut Pasteur international MLST database for L. monocytogenes](https://bigsdb.pasteur.fr/listeria/). Genetic comparison between isolates was performed by a core single nucleotide polymorphism (SNP) alignment for each clonal complex generated using SnapperDB (Dallman et al., 2018). Pairwise comparisons of SNP distances were performed between cultures within similar clonal complexes (Dallman et al., 2018). Isolates within a 10 SNP single linkage cluster were considered to come from a common point source, with each strain differing by no more than 10 SNPs from at least one other isolate in the cluster (Dallman et al., 2018). Genomic data from food, environmental and clinical isolates are stored in a customised database (UKHSA Gastro Data Warehouse). Sequence data is available through https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA248549.
To provide further characterisation of L. monocytogenes isolates including identification of AMR and virulence genes, isolates were sent to Quadram Institute Bioscience. Genomic Illumina reads were trimmed using Trimmomatic (Bolger et al., 2014). Trimmed genomic reads were assembled using Spades (Bankevich et al., 2012) in “careful” mode. The quality of the assemblies was assessed using QUAST (Gurevich et al., 2013), CheckM (Parks et al., 2015) and by aligning reads to the assemblies using the Burrows-Wheeler aligner (BWA) (Li & Durbin, 2009). Assemblies were accepted if they consisted of less than 500 contigs that were over 500 bp, less than 50 duplicate genes and had a mean read depth of the four largest contigs above 30.
AMR genes, virulence genes and plasmid replicons were identified using ARIBA (Hunt et al., 2017) and the ResFinder (Feldgarden et al., 2019), VFDB (Chen et al., 2016) and PlasmidFinder (Carattoli et al., 2014) databases, respectively. Metal-tolerance genes were identified genome assemblies using tBLASTn (Gertz et al., 2006) and the BacMet (Pal et al., 2014) database with 90% identity and coverage cut-offs.
Assembled genomes were annotated using BAKTA (Schwengers et al., 2021) and the genes were clustered using Roary (Page et al., 2015) using a 95% identity cut-off. RaXML (Stamatakis, 2014) was used to form a maximum likelihood tree using a GTR (Tavare, 1986) from the core gene alignment.
Figures were created using R (R core team, 2019).
For each sequence type (ST) where L. monocytogenes isolates from salmon and human cases were within a 10 SNP single linkage cluster, a reference genome was obtained: GCA_048319835.1 for ST8 and GCA_048631585.1 for ST121. PHASTEST (Wishart et al., 2023) was used to identify prophage regions in the reference genomes that were then blocked out, and plasmid contigs were removed. For these STs, Snippy (Hall et al., 2024) was used to align trimmed reads to the prophage removed reference genome before Gubbins (Croucher et al., 2015) was used to remove areas of recombination. RAxML was used to form maximum likelihood trees from the non-recombinant SNPs.
4.6. Characterisation of historical Listeria monocytogenes from previously tested raw salmon samples
L. monocytogenes isolated from food samples tested by public health laboratories in England, Scotland, Wales and Northern Ireland are routinely sent to UKHSA GBRU for WGS analysis, and all results as well as metadata are recorded on a database. All sequence data is made publicly available through a data repository known as the Sequence Read Archive (SRA).
To compare L. monocytogenes WGS results from this study of raw salmon to other raw salmon isolates detected over recent years, the UKHSA database was reviewed to identify representative L. monocytogenes isolates from raw salmon samples previously submitted to GBRU for WGS. The SRA succession numbers for 15 isolates that had been previously sent to GBRU were provided to Quadram Institute Bioscience for bioinformatic analysis.
Sequences were examined for the presence of genes encoding antimicrobial resistance and biocide resistance and virulence factors as described above.
4.7. Statistical analysis and data visualisation
Differences in microbial prevalence between retailers were investigating using the Chi-squared test in GraphPad (Dotmatics). Maps were created using RStudio 2024.04.2.
4.8. Quality assurance
All the salmon testing laboratories that participated in this study are UKAS-accredited to ISO 17025 and participate in External Quality Assurance (EQA) schemes. Laboratories carrying out MIC determination and further identification of strains also work within quality management systems (ISO 9001) and participate in EQA schemes. All analyses were performed by trained and competent staff.
5. Results
5.1. Description of salmon samples collected for microbiological examination
A total of 307 samples of raw salmon fillets were examined between January and December 2024. These included samples from 12 different approved establishments, with countries of origin recorded as Scotland (n=171; 56%), Norway (n=89; 29%) or a combination of Norway, Scotland and/or UK (n=47; 15%). Farm details were not available for the majority of samples (n=265; 86%) due to limited labelling information.
Samples were collected from outlets of nine major supermarket chains across all regions of England (East, East Midlands, London, Northeast, Northwest, Southeast, Southwest, West Midlands, and Yorkshire and the Humber), Scotland, Wales and Northern Ireland. Sampling locations are illustrated in Figure 1.
5.2. Detection of target organisms in samples of raw salmon
Generic E. coli were detected in 108 out of 307 salmon samples (35%; Table 3). The ‘detection method’ used here confirms that E. coli is present in a 25g portion of fish but does not provide an estimation of the count of E. coli present per gram of sample. Therefore, an enumeration technique was additionally used to identify those samples with a higher count of E. coli. The enumeration method has a lower detection limit of 20 cfu/g. Of all the samples in which E. coli was detected, the level of E. coli was less than 20 cfu/g in 107 samples, with the remaining sample having a level of 20 cfu/g.
Specific AMR E. coli targeted through selective culturing (i.e. ESBL-/AmpC-producing, carbapenem-resistant and colistin-resistant E. coli) were not detected in any samples. Samples where E. coli was detected were distributed across the UK (Figure 2).
L. monocytogenes was isolated from five out of 307 (1.6%) salmon fillet samples and were characterised as sequence types ST7 (clonal complex CC7), ST8 (CC8), ST18 (CC18), ST121 (CC121) and ST425 (CC90) respectively (all lineage II; serotype 1/2a). Listeria species other than L. monocytogenes were detected in 31 out of 307 samples (10%); these included L. welshimeri (5.9%, 18/307), L. innocua (3.9%, 12/307) or both L. innocua and L. welshimeri (0.3%, 1/307). As with E. coli, samples with L. monocytogenes and other Listeria species did not concentrate in any particular area of the UK (Figure 2).
The five L. monocytogenes strains were isolated from salmon samples that originated from four production codes (i.e. different processing establishments), with countries of origin being Scotland for three of the samples, Norway for one and one described as ‘Scotland and Norway’. Four were collected from the same supermarket chain (Retailer A; see Table 4) while one was collected from a separate supermarket, Retailer F.
There was a difference between the originating supermarket chains for samples in which either L. innocua or L. welshimeri were detected (Table 4). For example, L. innocua was found in >2% of samples from retailers D and G, whereas L. welshimeri was detected in >2% of samples from retailers A, B, C, E and F. The difference in L. welshimeri frequency between the different retailers was significant (Chi-squared test; p=0.007), while the difference for L. innocua was not significant (p=0.07). When the approved establishment codes were reviewed, L. innocua was detected in samples originating from three different establishments and L. welshimeri from six establishments. Two establishments were associated with both L. innocua- and L. welshimeri-contaminated samples.
5.3. Antimicrobial resistance of Listeria monocytogenes and E. coli isolates
Out of 108 E. coli isolates recovered in the survey, 102 were analysed using broth microdilution to determine their MIC (viable cultures of the remaining six isolates were not available for MIC determination). Of those, four (3.9%) were resistant to ampicillin and the remainder (98; 96.0%) were sensitive to all antimicrobials tested (ampicillin, amikacin, azithromycin, cefotaxime, ceftazidime, chloramphenicol, ciprofloxacin, colistin, gentamicin, meropenem, nalidixic acid, sulfonamides, tetracycline, tigecycline, trimethoprim).
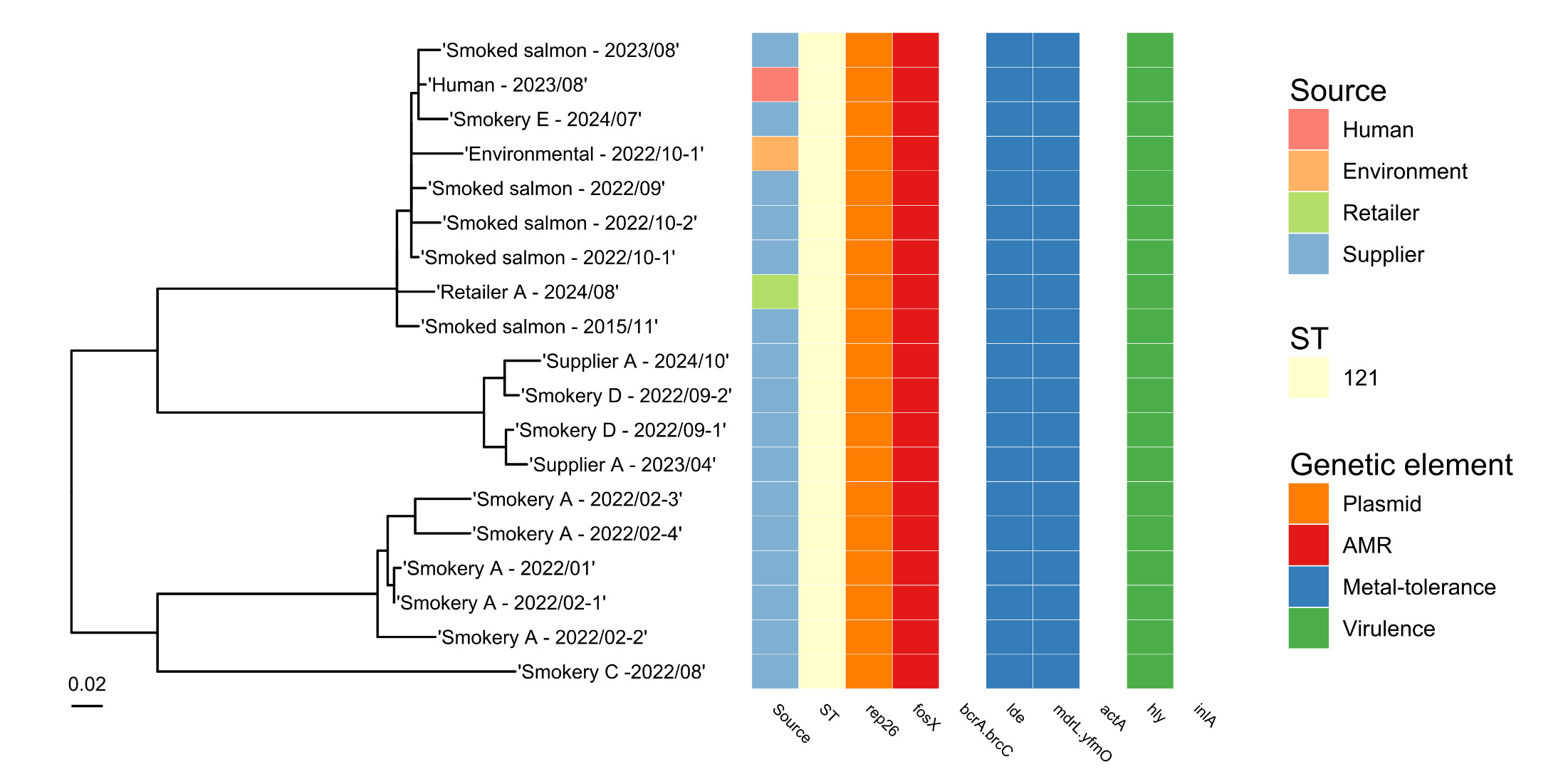
All five L. monocytogenes isolates possessed the fosX2 gene coding for fosfomycin resistance (Table 5) but no other AMR genes were detected during analysis of whole genome sequences for these strains. All isolates also carried the mdrL/yfmO gene that may be linked with resistance to the disinfectant, benzalkonium chloride (Figure 3). The ST121 strain contained the qacH gene that is associated with resistance to quaternary ammonium compounds (QACs) which are used as cleaning and disinfection agents in a variety of food businesses.
5.4. Phylogenetic links between Listeria monocytogenes from raw salmon samples and human cases
When the Whole Genome Sequence data was analysed for L. monocytogenes strains from retail salmon samples, one isolate (ST121) fell within a 10 SNP cluster with a human case of listeriosis that was detected in August 2023 (Figure 4). This was from a salmon sample collected from Retailer A in Scotland in August 2024. Also linked with the salmon and patient isolates at the 10 SNP level were isolates from 10 other foods (these samples were not collected for the purposes of the raw salmon study) and four environmental swabs tested by UKHSA and other labs between 2013 and 2024. Where further details were available for these isolates, the food samples were all raw or smoked salmon, and three of the environmental swabs were from a fish smokehouse.
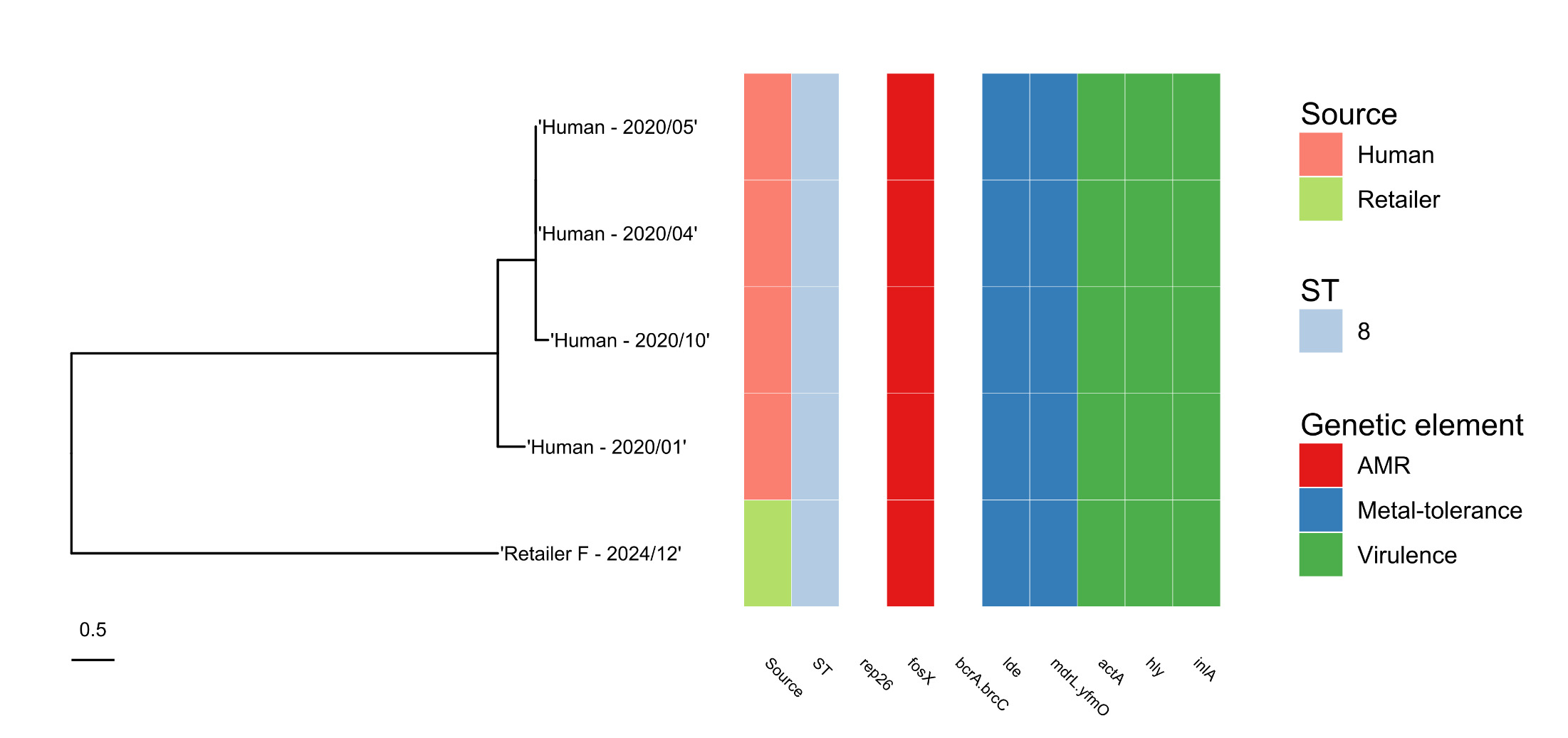
A second isolate (ST8), detected in a sample from an outlet of Retailer B located in Yorkshire in December 2024, fell into a 10 SNP cluster with four patients isolates (submitted to UKHSA GBRU between January and October 2020) as well as four food isolates submitted between 2015 and 2022 and a number of environmental isolates tested in 2021 (Figure 5). Of the food isolates falling within this cluster, three were from smoked salmon samples and one was from a sample of cheese.
The other three L. monocytogenes isolates were not linked with patient isolates available in the UKHSA GBRU database.
5.5. Characterisation of historical Listeria monocytogenes from previously tested raw salmon samples
Fifteen Whole Genome Sequences were analysed from L. monocytogenes isolates detected in raw salmon samples examined routinely in public health labs in England and Northern Ireland between 2018 and 2024. These were sampled from five fish smokery businesses, one fish wholesaler and two catering businesses, and included samples from businesses in London, East, Southeast, Southwest and Northwest regions of England as well as one site in Northern Ireland.
Strains were characterised as serotype 1/2a: ST121 (n=11), ST20 (n=1), ST204 (n=1), ST399 (n=1) and serotype 1/2b: ST87 (n=1). All ST121, ST20 and ST204 genomes contained the rep26 plasmid replicon, while no plasmid replicons were detected in the remaining strains.
All 15 L. monocytogenes isolates possessed the fosX2 gene coding for fosfomycin resistance (Table 6) but no other AMR genes were detected. All isolates also carried the lde and mdrL/yfmO genes. Two isolates (one ST20 and one ST204, collected from two different premises) also carried the bcrA and bcrC genes encoding resistance to benzalkonium chloride. In addition, all ST121 isolates contained the qacH gene that is associated with resistance to QACs, but this gene was not present in isolates belonging to other STs.
5.6. Identification of virulence genes in Listeria monocytogenes isolates from retail raw salmon and previously tested raw salmon samples
Amongst the 20 L. monocytogenes genomes, 35 virulence genes were identified (as defined in the Virulence Finder Database; Chen et al., 2016). Of these, 22 were found in all genomes (i.e. core virulence genes), whilst 13 varied in their presence amongst the genomes.
All L. monocytogenes isolates contained the hly gene that encodes for Listeriolysin O, essential for L. monocytogenes virulence (Cossart et al., 1989), but ST121 isolates did not have a functional actA gene (required for re-organising actin in eukaryotic cells; Pistor et al., 1994) or inlA gene (required for crossing the intestinal epithelial barrier; Pentecost et al., 2006).
6. Discussion
This study of raw, chilled, prepacked farmed salmon fillets collected from supermarkets in the UK demonstrated a low prevalence of L. monocytogenes in this commodity, as well as low levels of both generic E. coli and AMR E. coli. This provides reassurance for consumers and regulators of the generally low microbiological risk associated with raw salmon on the market. This risk can be reduced even further through hygienic handling and appropriate cooking of salmon by consumers.
Listeria monocytogenes occurs naturally in the terrestrial and aquatic environments, being found in both fresh and salt water and in foods associated with these environments. Since this organism can survive and grow at refrigeration temperatures, it may persist and out-compete other bacterial contaminants in raw ingredients from these environments (Jami et al., 2014). Recent investigations of L. monocytogenes contamination of smoked salmon, and associated cases of illness, have highlighted the risk posed by contaminated smoked salmon to vulnerable consumers (ECDC-EFSA, 2024b, 2024a). Between 2017 and 2019, there were seven recalls of smoked salmon and slightly salted salmon in the United States as a result of concerns over L. monocytogenes contamination (FDA, 2020). Furthermore, a study of 785 smoked fish samples in England in 2022/2023 found L. monocytogenes in 3.6% of samples overall, with 6.4% of cold smoked salmon samples being contaminated with this organism (UKHSA, unpublished data). In contrast, 1.6% of raw salmon fillet samples examined were contaminated with L. monocytogenes in the study reported here. While there were some differences in study design between the UKHSA study of smoked fish and this study of raw salmon (i.e. the UKHSA smoked fish samples were collected largely from retail (89%) but with some samples from catering and manufacturing settings as well; the smoked fish products were not specifically chosen to be fillets but represented any cuts that were available at the time of sampling), the difference in L. monocytogenes contamination rates in the two studies indicates that the presence of L. monocytogenes in the smoked products may predominantly be a result of contamination from within the smokehouse and/or processing plant, rather than simply representing the bacteria present in the raw fish prior to smoking. This hypothesis is backed up by a meta-analysis of results for 14,496 samples of fish, fish products and fish processing environments, reported in publications from 2013 to 2023, which found that the pooled prevalence of L. monocytogenes and Listeria species in raw fish was 5.8% and 12.2% respectively (Zakrzewski et al., 2024). In comparison, prevalence in ready-to-eat (RTE) products was higher, with L. monocytogenes in 14.5% and Listeria species in 21.7% of samples.
The ability of L. monocytogenes to establish biofilms and persist in the processing environment (Fagerlund et al., 2022; Lee et al., 2019) as well as its ability to multiply at refrigeration temperatures help to explain why RTE fish products may become more contaminated with this pathogen compared to the raw fish. The ability of these bacteria to develop resistance to biocides also contributes to their survival in the food processing environment (Duze et al., 2021).
Listeria species other than L. monocytogenes (namely L. innocua and L. welshimeri) were detected in 10% of raw salmon samples examined as part of this study. In general, Listeria species other than L. monocytogenes are not considered to be pathogenic but can be an indicator of conditions that could allow contamination and/or growth of L. monocytogenes. Each of these species appeared to be more commonly associated with certain retailers, with different approved establishments being more frequently associated with each (apart from two establishments linked to both L. innocua and L. welshimeri contamination). This may indicate the presence of persistent contamination of one or both species within the processing environment of the different approved establishments that each supply different retailers.
Acquired antimicrobial resistance genes were not detected in any of the 20 L. monocytogenes strains, with the fosX2 gene (encoding fosfomycin resistance) being the only AMR gene detected. The fosX2 gene is present in the core genome of L. monocytogenes, conferring intrinsic fosfomycin resistance to all L. monocytogenes strains (i.e. naturally occurring resistance that is not dependent on mutation or gain of further genes). However, studies have shown that the effect of fosfomycin is greater in vivo than it appears in vitro, due to the presence of other genes (hpt and prfA) that enable increased fosfomycin uptake into the bacterial cell (Scortti et al., 2018). There was evidence of hpt and prfA genes in all the raw salmon isolates examined in this study.
The absence of other AMR genes in the isolates examined during the current study indicates that resistance to the main antimicrobials used in the treatment of listeriosis, such as ampicillin and gentamicin, is low in salmon products in the UK. The absence of resistance to tetracycline is also of interest, given that the UK Veterinary Antibiotic Resistance and Sales Surveillance (VARSS) report for 2021 showed an increase in usage of antimicrobials in salmon farming between 2017 and 2021, with oxytetracyline accounting for 86% of total antimicrobial use in 2021 (UK-VARSS 2021, 2022). However, given that L. monocytogenes strains in the salmon samples may have been introduced during processing rather than originating from the aquaculture environment, perhaps it is unsurprising that antimicrobial usage during farming is not reflected in tetracycline resistance amongst the salmon isolates.
A study of 5,339 L. monocytogenes isolates from clinical and food samples in France (Moura et al., 2024) also showed low levels of resistance to clinically important antimicrobials, with acquired resistance detected in 2.2% of isolates. This included resistance to tetracyclines, trimethoprim, lincosamides and macrolides but not the preferred treatment options, ampicillin or gentamicin. Furthermore, Moura et al. (2024) found that acquired resistance was more prevalent in food than clinical isolates, and more prevalent in isolates that also showed disinfectant or stress resistance.
Genes conferring potential resistance to quaternary ammonium compounds (QACs) that are used as disinfectants in the food industry were detected in all of the L. monocytogenes isolates examined during this study. Resistance to these disinfectants contributes to the ability of these strains to persist in the food production environment, and confers a survival advantage over sensitive strains (Martínez-Suárez et al., 2016). This may include the ability to form biofilms on surfaces in the presence of biocide, particularly in niches where increased moisture or organic matter result in a reduced concentration of biocide, or where a residual lower concentration of disinfectant remains on surfaces after cleaning and rinsing (Møretrø et al., 2017).
All the L. monocytogenes isolates from raw salmon in this study belonged to lineage II. Lineage II strains tend to be commonly found in foods and the environment, as well as being associated with sporadic cases of human infection (Orsi et al., 2011). In contrast, most outbreaks of listeriosis in humans are caused by strains belonging to lineage I. The majority of L. monocytogenes isolates from raw salmon (both the retail study and the isolates from supply and catering premises) were ST121, which is recognised to be well adapted to persist in the food production environment, but is characterised as ‘hypovirulent’ (i.e. less likely to cause illness) due to its truncated inlA gene, which affects its ability to cross the intestinal epithelial barrier (Maury et al., 2016; Rychli et al., 2017). Despite this reduced virulence, an ST121 isolate from a retail raw salmon sample was closely related to an isolate from a human case of listeriosis (less than 10 SNPs difference between salmon and human isolates). An ST8 salmon isolate was also related to four isolates from human cases of infection at the 10 SNP level. This strain had a fully functional inlA gene.
The plasmid replicon, ‘rep26’, was detected in 14 of the 20 L. monocytogenes strains analysed. The identification of different replicon types allows an understanding of genetic diversity of plasmids in L. monocytogenes populations. This is important in understanding the spread of antibiotic resistance and/or other virulence factors carried by these plasmids. It has been shown that plasmids are found more frequently in L. monocytogenes isolates from food and environmental samples than from clinical isolates (Kuenne et al., 2010). Moreover, those strains persisting in the food processing environment (i.e. recovered from the same environment on successive sampling occasions) showed the presence of plasmids more frequently than sporadic strains that were only detected on a single occasion (Harvey & Gilmour, 2001).
There is limited data available in published literature regarding the prevalence of AMR E. coli in salmon. However, where studies have been published, prevalence of ESBL-producing E. coli in raw fish generally appears to be relatively low. For example, ESBL E. coli were detected in only one of 30 raw retail salmon samples in Portugal (Silva et al., 2019) and in one of 28 raw salmon samples on retail sale in Spain (Vitas AI et al., 2018). A study of seafood on retail sale in the United States, including 710 salmon samples (as well as shrimp and tilapia), also demonstrated pan-susceptibility in Salmonella and Pseudomonas aeruginosa and low prevalence of resistance (<10%) to the majority of antimicrobials tested for Aeromonas, Vibrio, Staphylococcus and Enterococcus species (Tate et al., 2022). A study of rainbow trout in Lebanon demonstrated the presence of the mcr-1 gene, which confers colistin resistance, in E. coli isolates from fish guts (Hassan et al., 2020); analysis of these isolates by WGS indicated that the mcr-1 gene was present on transmissible plasmids, demonstrating the potential for the dissemination of colistin resistance genes in the aquatic environment. A risk profile of carbapenem-resistant E. coli (CRE) in shrimp and salmon on retail sale in Canada concluded that data on CRE occurrence and distribution in seafood are limited and that periodic surveillance should be implemented due to the critically important nature of the carbapenem class of antimicrobials (Loest et al., 2022). Results from the current study were encouraging in that antimicrobial resistance in E. coli appeared to be low, with only ampicillin resistance detected in 3.9% of E. coli isolates and no detection of ESBL-producing, carbapenemase-producing or colistin-resistant E. coli in any of the raw salmon samples examined. As with the L. monocytogenes isolates, the lack of resistance to tetracycline in E. coli was reassuring, given the significant quantities of oxytetracycline used in salmon farming in the UK (UK-VARSS 2021, 2022). However, it must be noted that E. coli detected in the salmon fillets may have been introduced during the post-harvest transport and/or processing stages rather than during aquaculture, and therefore, it is not possible to determine a direct link (or otherwise) between antibiotic usage in the salmon farms and the AMR results seen in this study.
The majority of salmon samples tested under this survey originated from Scotland (UK) and Norway. The UK Veterinary Medicines Directorate (UK-VARSS 2023, 2024) reported a 7% increase in antibiotic active ingredient usage by the UK salmon industry in 2023 compared to 2022, and a 24% increase since 2017. This usage comprised 50% oxytetracycline and 50% florfenicol, which represents an overall reduction in oxytetracycline since 2017 but a 3.5-fold increase in florfenicol usage. A statement from ‘Salmon Scotland’ clarified that antibiotics are only used in the salmon industry in response to clinical evidence of bacterial infections. The usage of antimicrobials in humans and animals in the EU, Norway and Iceland is reported by the Joint Inter-Agency Antimicrobial Consumption and Resistance Analysis (JIACRA) group. Their fourth report, covering the period from 2019 to 2021 (JIACRA, 2022 Antimicrobial consumption and resistance in bacteria from humans and food-producing animals) indicates that consumption of antimicrobials in Norway for use in food-producing animals is very low in general. However, no specific data was available on aquaculture usage.
Overall, it appears that antimicrobial usage in salmon production is lower than in the production of many other food-producing animals, which may at least partially explain the relatively lower rates of AMR in the samples tested as part of this study compared to similar studies of other food products of animal origin. For example, a study of frozen breaded chicken products on retail sale in the UK in 2021 found ESBL-producing E. coli in 4.8% of samples (Willis et al., 2023), while earlier studies of fresh chicken in the UK found higher levels of ESBL-E. coli: 10.1% in 2017 (Willis et al., 2018), 8.4% in 2018 (Randall et al., 2020) and 12.4% in 2020 (APHA, 2021). A survey of 105 beef and 105 pork samples on retail sale in the UK in 2021 resulted in the isolation of ESBL-E. coli from 0.95% of beef and 3.8% of pork samples (APHA, 2022).
This study of 307 samples of raw salmon fillets has provided reassurance that the microbiological risk to UK consumers from L. monocytogenes and AMR in retail salmon is currently low. However, it is important to continue to monitor microbiological risks in seafood, as the potential for antimicrobial usage in worldwide aquaculture to encourage the development of AMR in these products is well recognised. While the prevalence of L. monocytogenes in raw salmon samples was also low, the ability of this pathogen to persist in processing environments highlights the need for consumers and food businesses to be well-informed about safe handling practices for raw and processed salmon.
According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST), a microorganism is defined as wild type for a species by the absence of acquired and mutational resistance mechanisms to the drug in question (see ACMSF AMR Working Groups paper on AMR terminology).
ECOFFs are determined by a different approach than clinical breakpoints (CBPs), and do not take into account the results of clinical efficacy studies, dosing and route of administration of the antimicrobial agents, nor the drug’s pharmacokinetic and pharmacodynamic parameters in humans or animal species in which such substances are used (see ACMSF AMR Working Groups paper on AMR terminology).


_*l._monocytogenes*__(b)_other_*lis.png)



_*l._monocytogenes*__(b)_other_*lis.png)