Executive Summary
Whole-genome sequencing (WGS) for foodborne disease (FBD) surveillance provides many benefits, including new insights in disease transmission, virulence and antimicrobial resistance (AMR), fast and precise outbreak tracing and source attribution, as well as, enabling streamlined and reproducible analysis through digital data that from a technical point of view can be easily shared. The National foodborne disease genomic data platform, developed as part of the PATH-SAFE programme, will offer a trusted environment for WGS data sharing and analysis for UK agencies involved in FBD surveillance. The platform has initially been built for Salmonella with the intention to expand it to other organisms later. Where possible, the platform has drawn on existing and validated solutions as implemented in EnteroBase, PubMLST and Pathogenwatch. The platform is hosted on CLIMB-BIG-DATA and has been built to enable interoperability between analytical tools and databases. Although a fully interoperable bioinformatics system for FBD and AMR surveillance is not practically feasible at this time, it should be a long-term goal.
Recommendations on which tools to use for molecular surveillance of Salmonella have been developed in consultation with Community Input Advisory Groups (CIAGs) on 1) technical aspects of FBD surveillance, 2) AMR risk determinants, 3) data standards for FBD surveillance, 4) considerations for international molecular FBD surveillance.
The purpose of this document is to act as a standard reference for methodologies for genomic surveillance of FBD to support FSA goals and achieve the benefits laid out above. The recommendations have been developed using non-typhoidal Salmonella as an example.
Contributors
Abbreviations
1. Background & objectives
Foodborne diseases (FBDs) are an increasing threat to public health and food security. The World Health Organization (WHO) estimates that 600 million episodes of ill health and 420,000 deaths are caused by FBDs annually (World Health Organization, 2024). In the UK foodborne pathogens cause 2.4 million cases of disease and a cost of £9.1 billion annually (Food Standards Agency, n.d.).
Whole-genome sequencing (WGS) provides a vast amount of information and the highest possible resolution for pathogen typing. The application of WGS in FBD surveillance can provide public health agencies with information on the source and extent of outbreaks, the emergence and spread of antimicrobial resistance (AMR) and other risk factors, and support policy development and decisions on interventions for FBD control.
Benefits of WGS for FBD surveillance include:
-
New insights in disease transmission and virulence when combined with epidemiological, clinical, veterinary and microbiological metadata, enabling more rigorous risk assessment;
-
Subtyping of pathogens for surveillance and outbreak investigation;
-
Fast and precise analysis which improves outbreak tracing and source attribution and can lead to reduced outbreak size and potential cost savings by speeding up time to effective response;
-
Genome-wide, high-resolution typing of AMR pathogens which can aid targeted response and our understanding of AMR evolution;
-
Differentiation between modes of AMR transmission, including via mobile genetic elements, e.g. plasmids;
-
Digital data facilitate reproducible analyses and improve comparability between different laboratories;
-
Streamlined technology that can test for multiple pathogens or resistance mechanisms at once at no additional cost;
-
Data storage in comprehensive genome databases which facilitates data sharing and application of machine learning tools for analysis.
WGS data alone does not provide actionable insights to public health and food security agencies. Rather, WGS data need to be combined with epidemiological, clinical, veterinary and microbiological metadata to inform decision-making and interventions for foodborne pathogens. WGS data require a high degree of standardisation for their benefits to be maximised. The need for data standardisation is particularly pertinent in the food safety space because multiple agencies are involved in surveillance, all independently generating data. Standardisation supports interoperability of data and quality assurance. At the same time, the greater resolution of WGS data compared to phenotypic surveillance data requires stringent data protection regulations, especially when WGS data are combined with sensitive metadata.
In light of the increasing importance of genomic surveillance of FBD, the UK Food Standards Agency (FSA) has commissioned a comprehensive piece of work to build a high-performing, user-friendly and flexible platform for genomic surveillance of FBD (“National foodborne disease genomic data platform”, also referred to as “the PATH-SAFE platform” or “the platform” in the following). As a minimum, the platform will support the use cases for WGS surveillance of FBD in public health and food security listed in Table 1.
The platform is meant to facilitate data sharing and interoperability between the various agencies involved in FBD surveillance in the UK. These agencies are represented in the End User Group of the PATH-SAFE consortium and include the FSA, FSS, APHA, UKHSA and PHS. The platform has drawn on existing tools and solutions to avoid duplication of efforts and to ensure that all components are validated and of proven utility. Recommendations on which tools to use for molecular surveillance of Salmonella have been developed in consultation with Community Input Advisory Groups (CIAGs) on 1) technical aspects of FBD surveillance, 2) AMR risk determinants, 3) data standards for FBD surveillance, 4) considerations for international molecular FBD surveillance.
The purpose of this document is to act as a standard reference for methodologies for genomic surveillance of FBD to support FSA goals and achieve the benefits laid out above. The recommendations have been developed using non-typhoidal Salmonella as an example. Typhoidal Salmonella has been excluded from initial considerations because it is not normally a risk in food production and supply chains. Using Salmonella as a test case for the new PATH-SAFE platform has the advantage that serotypes are consistent with evolutionary groups which facilitates the use, and backward compatibility, of novel molecular surveillance tools with classic microbiological methods. Moreover, tools for antibiogram prediction including classification of susceptible/intermediate/resistant clones with regards to specific antimicrobial drugs exist and have been validated for Salmonella (Sherry et al., 2023). Tools and recommendations for other foodborne pathogens will be added to the platform at a later stage. The processing, analysis and reporting steps and underlying technical components of the platform for WGS surveillance in FBD and how they relate to sections in this document is illustrated in Figure 1.
The platform is hosted on CLIMB-BIG-DATA (Cloud Infrastructure for Big Data Microbial Bioinformatics, https://www.climb.ac.uk/), the base platform also used by COG-UK (COVID-19 Genome Consortium United Kingdom). CLIMB-BIG-DATA offers secure, scalable hosting and containerised analytics pipeline management for decentralised sequencing and centralised data analysis. The platform will assemble genomes via the CLIMB infrastructure.
The PATH-SAFE platform draws on EnteroBase (Zhou et al., 2020), an integrated software environment for analysing and visualising genomic variation within enteric bacteria, for its assembly pipeline. It was explored whether genomic data uploaded to the PATH-SAFE platform could be fed into EnteroBase for use by the FBD WGS surveillance community, due to data sharing sensitivities this was not implemented. EnteroBase maintains the HierCC clustering scheme that will be utilised by the platform (see Section 2).
In addition, the platform utilises analysis schemes implemented in PubMLST (https://pubmlst.org/), a public database for molecular typing and the analysis of microbial genome diversity, for gold standard Salmonella typing (MLST, cgMLST).
A key component of the PATH-SAFE platform is Pathogenwatch (https://pathogen.watch/), a platform for genomic surveillance, providing analysis tools, including AMR and risk marker prediction, clustering analysis and reporting. The user interface of the platform is provided by the Pathogenwatch infrastructure.
2. Technical aspects of genomic surveillance of foodborne diseases
This section covers the technical requirements of using WGS for FBD surveillance. The recommendations in this section are not necessarily about whether one tool is better than another, but rather on how tools support the interpretation of molecular surveillance data and how they can help analysts meet technical and legislative requirements. The recommendations are deliberately not prescriptive but allow for the use of multiple validated and comparable solutions alongside each other. Some of the recommendations are use case-specific or linked to critical control points in the food supply chain.
In the following subsections, the document lays out technical requirements and best practice analytics for genomic surveillance of FBD for the above mentioned use cases. The recommendations will ensure flexibility of the platform with a view to future scale-up and technological and research advances, for example, increased use of long-read and hybrid sequencing platforms.
Validation reports for functional tools included as part of the PATH-SAFE platform are included in Appendix 4.
2.1. Assembly pipeline and quality control metrics
An accurate high-quality assembly is the prerequisite for downstream analysis steps, such as AMR detection (covered in detail in Section 3) or clustering of sequences. As part of the PATH-SAFE consortium, FSA commissioned the University of Warwick Bioinformatics Research Technology Platform to evaluate and compare different assemblers and assembly pipelines for Salmonella (EnteroBase, 2023) [reference report]. The main recommendations from this evaluation are summarised in the following.
The evaluation included ten assemblers suitable for Illumina short-read sequences of enteric bacterial genomes and compared the quality of assembled genomes. Important quality metrics capture the number and length of assembled contigs, the number of called bases, genome completeness, the proportion of failed assemblies and the variance of these metrics when repeatedly assembling the same test data. There were few differences between the tested assemblers, apart from Velvet (EnteroBase, 2023; Zerbino & Birney, 2008) which produced assemblies of inconsistent quality and therefore should not be used for assembling Salmonella genomes.
The primary recommendation from the evaluation report is to use a SPAdes based assembler (Bankevich et al., 2012; EnteroBase, 2023) for Salmonella genome assembly because it consistently produces high-quality assemblies and can process long-read as well as short-read sequence data (both long-read only and hybrid long-read/short-read sequences).
Different assembly pipelines offer pre- and post-processing steps that can improve the quality of genome assemblies. The evaluation of different assembly pipelines concluded that adapter trimming, trimming of low-quality sequences, down-sampling of very high coverage reads and post-assembly polishing steps (processing of a draft assembly to remove artefacts and improve local and overall consensus accuracy) slightly but consistently improved assembly quality. Consequently, these functionalities should be implemented in the preferred assembly pipeline. The EToKi (Zhou, 2023), Pathogenwatch (Argimón et al., 2021) and Unicycler (Wick et al., 2017) pipelines provide the relevant functionalities in conjunction with SPAdes for the assembly step, and offer comparable high quality of assembled genomes.
The assembly pipeline for the PATH-SAFE platform should have the capacity to work with hybrid short- and long-read sequences and with long-read only data, as long-read sequences are increasingly generated by novel sequencing technologies, and there is the possibility that sequencing will move away from generating short-read data. EToKi and Unicycler have the capability to process long-read sequence data. Therefore, using one of these pipelines will increase future comparability of generated genome assemblies. Since EToKi is already used by EnteroBase, which will feed into the platform, the recommendation is to use the EToKi pipeline as the foundation for the PATH-SAFE genome assemblies.
Recommendations:
-
Genomes assembled by SPAdes, as implemented in pipelines that incorporate suitable pre- and post-processing steps, including EToKi, Unicycler or Pathogenwatch, could all be accepted for within the PATH-SAFE platform.
-
The EToKi pipeline based on the SPAdes assembler and key pre- and post-processing steps is used by EnteroBase, and has been shown to consistently produce good quality genome assemblies. It is therefore recommended as the assembly pipeline implemented within the PATH-SAFE data platform.
-
Velvet produced assemblies of inconsistent quality and should not be used for assembling Salmonella genomes.
2.2. Analysis framework
Important functional requirements for molecular surveillance of foodborne pathogens include typing and clustering. Typing is important to characterise isolates and to understand which lineages are involved in disease outbreaks. Clustering is important to understand how isolates and lineages relate to each other.
Molecular typing methods are pathogen-specific and for Salmonella include MLST (multi-locus sequence typing) (Achtman et al., 2012), cgMLST (core genome multi-locus sequence typing) (Pearce et al., 2018), wgMLST (whole genome multi-locus sequence typing) (Leeper et al., 2023), SNP-based typing (Pearce et al., 2018) and in silico serotyping (Uelze et al., 2020). Of these methods, MLST and cgMLST are the most widely used because they rely on relatively small sets of defined genes. wgMLST, which analyses a larger number of allelic loci incurs a greater computational overhead than cgMLST, lacks stringent validation compared to other typing methods, and currently does not add value for routine molecular surveillance activities (Leeper et al., 2023; Mohammed & Thapa, 2020). However, wgMLST is implemented in EnteroBase and may be used as an investigational tool to provide additional information during an FBD outbreak. Previous studies have shown that cgMLST-based and SNP-based typing deliver concordant results (Chattaway et al., 2023).
The reliability of cgMLST typing depends on the quality of assembled genomes, in particular, on QC thresholds for missing data. Therefore, validation of tools for cgMLST should include genomes that are known to be incomplete to ensure that the tools will perform adequately in real-world situations (e.g. FBD outbreaks).
In silico serotyping uses genetic sequence data to determine which serotype a Salmonella isolate belongs to. Historically, serotyping has been the major phenotypic subtyping method for Salmonella isolates and nearly 2500 serotypes are defined (Grimont & Weill, 2007). Accurate in silico serotyping enables backward compatibility with historic databases. Moreover, Salmonella serotype determination is required by some agencies as part of surveillance, e.g. APHA. Two widely used, validated in silico serotyping tools for Salmonella are SISTR (Yoshida et al., 2016) and SeqSero2 (Zhang et al., 2019). Both tools have a reported global prediction accuracy of more than 94% but may perform worse for rare serotypes, and SeqSero2 is less accurate for GC-biased sequences (Uelze et al., 2020; Yoshida et al., 2016; Zhang et al., 2019). As these tools have different strengths and weaknesses and are preferred by different agencies, the recommendation is that both tools are implemented in the PATH-SAFE platform.
Clustering and phylogenetic analyses are used to place the sequence of an isolate in its genetic context and to reconstruct transmission chains. Salmonella isolates can be clustered based on their MLST/cgMLST profiles. Tools for this task are implemented in EnteroBase and Pathogenwatch and as part of the COREugate pipeline (Achtman et al., 2022; Argimón et al., 2021; Higgs et al., 2022). Phylogenetic trees based on core SNP profile can achieve a higher resolution which can be useful, for example, to examine how human and animal isolates are related. However, trees are only tractable for a relatively small number of sequences that are known to be closely related, for example, because they fall into the same cluster. The PATH-SAFE platform will implement clustering based on cgMLST profiles. Clusters will be named according to their corresponding HierCC codes (assigned via EnteroBase). HierCC is a scalable clustering scheme based on cgMLST profiles that allows incremental, static, multi-level cluster assignment of genomes (Zhou et al., 2021).
Ideally, typing and clustering/phylogenetic tools should be able to correctly flag novel sequences that require further investigation, for example, sequences that cannot be typed or that do not neatly fall into a particular cluster.
Recommendations:
-
Tools for MLST and cgMLST should be available on the PATH-SAFE platform. wgMLST tools should be utilised for additional investigational use.
-
Validation datasets for cgMLST tools should include genomes known to be incomplete, as missing data are known to impact performance of these tools.
-
Both SISTR and SeqSero2 should be provided for molecular serotyping so that agencies can use their preferred tool and outputs can be compared.
-
The platform should implement tools for clustering based on MLST/cgMLST profiles. Clusters should be named according to their HierCC codes. Phylogenetic tree reconstruction tools based on SNP profile can be implemented for investigational use.
-
Typing and clustering tools should be able to flag novel sequences to trigger further investigation.
2.3. Integration and interoperability between platforms
Many of the analysis-specific tools discussed in the previous two subsections have been tested and validated and are in widespread use (Achtman et al., 2012; Argimón et al., 2021; Bankevich et al., 2012; Leeper et al., 2023; Pearce et al., 2018; Wick et al., 2017; Yoshida et al., 2016; Zhang et al., 2019; Zhou, 2023; Zhou et al., 2021). The main challenge will be to ensure interoperability between tools and platforms. In the first version of the PATH-SAFE platform, open-source analysis tools implemented individually or in other platforms are copied to the new platform and then used as internal tools. Ideally, a future feature of the platform would be to use API calls to analyses implemented in EnteroBase and other platforms, either to compare the results of the same analysis conducted by different tools (benchmarking) or to conduct additional analyses that are not implemented in the PATH-SAFE data platform. In this way both systems could use the same tool, instead of both systems maintaining and aligning two versions of the same tool, as is currently done. Although this functionality would be useful and a step to an open interoperable bioinformatics system for molecular surveillance of foodborne pathogens and AMR, more work is necessary to ensure the stability and security of such a system. Therefore, this functionality was not included in the first iteration of the PATH-SAFE platform.
Recommendations:
-
Where possible, the platform should rely on existing tools that are in widespread use and have already been validated.
-
The platform should be built to enable interoperability between tools and platforms. Although a fully interoperable bioinformatics system for FBD and AMR surveillance is not practically feasible at this time, it should be a long-term goal.
3. Genotypic markers of antimicrobial resistance in foodborne diseases
AMR surveillance largely relies on microbiological phenotypic methods that can be performed quickly using standard operating protocols for which laboratories can be certified. However, phenotypic methods cannot determine the mechanisms underlying resistance. WGS data can be used to identify the presence or absence of known resistance determinants and genetic risk factors, which serve as useful predictors of resistance phenotypes and importantly also as epidemiological markers. Notably, while WGS data cannot detect novel mechanisms of resistance that have not been observed before, they can easily be re-interrogated retrospectively for newly described markers.
To use genomics for detecting AMR and risk markers as part of FBD surveillance, consensus databases of AMR and risk markers and efficient bioinformatic tools are needed. This section defines best practices and collaborative mechanisms to ensure that all relevant genetic AMR and risk markers for Salmonella will be flagged by the PATH-SAFE platform. The concordance between phenotypic and genotypic resistance profiles is high but imperfect, because AMR genetic mechanisms are not always fully understood. However, the primary purpose of detecting AMR markers from WGS in the PATH-SAFE platform is neither to predict phenotypic resistance, nor to replace phenotypic testing, but to add information on AMR determinants. This data can provide information on the evolution and spread of novel AMR variants and facilitate tracking of mobile AMR genes between pathogen lineages and through the food supply chain.
As different sectors use different phenotyping methods and monitor for resistance to different drugs, genomics can play a unique role in linking these sectors and thus contribute to a unified surveillance system.
3.1. Drugs and risk markers to monitor
Both AMR genes and point mutations linked to resistance to specific drugs or drug classes should be monitored so that platform output can be organised in the style of an antibiogram. However, it is important to note that the PATH-SAFE platform AMR functionality will be for risk assessment at the population level and not for clinical decision-making at the individual level.
All resistance makers are potentially useful, regardless of the clinical relevance of the associated drug or phenotype, as they can have epidemiological value in defining and identifying specific outbreak strains and thus aid in communications with international partners who do not routinely sequence isolates. For example, the Salmonella Typhimurium strain behind the 2022 Kinder Surprise outbreak that affected multiple countries in Europe, including the UK, was characterised by an unusual AMR pattern that had not previously been seen in other UK isolates. This unusual AMR pattern could be used to link the strain to isolates from other affected countries including those that had not sequenced isolates (Larkin et al., 2022).
The exception in the context of Salmonella enterica is the aminoglycoside acetyltransferase aac(6’)-laa chromosomal core gene, which is hence the wildtype and so not a marker for risk in Salmonella. As such it has a status of ‘non-reported’ in the NCBI RefGenes database used by AMRFinderPlus and should not be reported as a resistance marker for Salmonella in the PATH-SAFE platform.
Potential sources of AMR markers are the NCBI Pathogen Detection Reference Gene Catalog (RefGenes) (National Institute of Health (US), 2024c), the ResFinder database (DTU Food, 2022) and the CARD database (Alcock et al., 2023). The ResFinder and RefGenes databases contain almost the same markers, with a few markers unique to each (Table 2). The reasons for the differences between the two databases are unclear, as both databases are being actively updated and maintained. ResFinder is curated by biologists reviewing published articles and collecting user communications on novel AMR genes or mutations that fulfil inclusion criteria into the ResFinder database (AMR genes) or PointFinder database (AMR mutations). Bioinformaticians then update the database and test it against the new entries (Florensa et al., 2022). Methods for curating the RefGenes database have been described in a recent paper (Feldgarden et al., 2022). In short, NCBI collects new AMR genes or mutations through literature searches, interorganisational data exchanges and collaborator requests. Inclusion of novel AMR genes, alleles or point mutations in RefGenes, requires experimental evidence or very close similarity to an experimentally verified protein. The CARD database includes a similar set of acquired genes and point mutations, but it also includes many intrinsic (core) genes. As a consequence, output generated by screening against the CARD database can be dense and confusing if all AMR genes and point mutations are queried.
Dictionaries mapping individual markers to specific drugs and drug classes are important for reporting. In particular, they are required to organise markers in the form of an antibiogram, which is generally accepted as the most interpretable and desirable format for users. This should be factored into the choice and implementation of the marker database for the PATH-SAFE platform. ResFinder maps genes to drug classes and specific antimicrobials. NCBI RefGenes maps genes to drug classes and subclasses, where a subclass is sometimes a drug class and sometimes a specific antimicrobial. This is mainly an issue for beta-lactam based antibiotics as the classes are quite broad (aztreonam, beta-lactam, carbapenem, cephalosporin, cephalothin, methicillin). Moreover, some aminoglycoside associated genes are differentiated into ‘subclass’ as combinations of specific drugs such as amikacin, gentamicin, kanamycin, tobramycin, spectinomycin, streptomycin, apramycin, neomycin, paromomycin; whereas other genes are assigned to subclass ‘aminoglycoside’ without further differentiation. This complicates use of RefGenes class/subclass for reporting. CARD currently maps markers to ontology giving broad drug classes. A database mapping to specific drugs is under development and should be considered in due course for future development of the PATH-SAFE platform.
Interoperability with international agencies needs to be considered. Otherwise, there is potential for confusion, e.g. due to differences in nomenclature of genes and mutations, or markers that are present in one database and not another. EU surveillance programmes currently utilise the ResFinder database, while US agencies utilise NCBI RefGenes (see Section 5). If, for example, an international agency relying on the NCBI RefGenes database identified an outbreak strain X containing marker Y, but Y is not included in the ResFinder database, an agency relying on the ResFinder database may find it difficult to recognise why Y is not reported and that their genomes match the outbreak strain.
Recommendations:
-
The platform should include all AMR markers in the source databases, regardless of clinical relevance of the associated drug or phenotype, as they can have epidemiological value in defining and identifying specific outbreak strains.
-
The exception is the Salmonella enterica core chromosomal gene aac(6’)-Iaa, which is included in the ResFinder database but should not be reported in the platform as this represents a wildtype genotype not conferring resistance. As the platform is expanded to other organisms, similar consideration will need to be given to organism-specific interpretation of AMR markers.
-
The platform should report AMR markers organised in the form of antibiograms, i.e. grouped by antimicrobial rather than class as far as possible, in order to be comparable to phenotype data.
-
Based on the current status of the source database(s), this is easiest to achieve using ResFinder as it includes a dictionary mapping markers to individual antimicrobials.
-
Notwithstanding this, the platform should implement solutions to allow interoperability with agencies using AMRFinderPlus. Options could be: (i) include acquired genes and point mutations that are present in either ResFinder or NCBI (i.e. combine the databases); (ii) implement both tools and allow users to choose which set to query. Each option adds complexities for reporting and versioning, and (i) would require manual curation of the dictionary mapping markers to antimicrobials.
-
The platform should be updated regularly to include updates to the source database(s), and record and report versions of the source database(s) included or utilised in each version of the platform software.
-
The platform should monitor changes to the content and inclusion criteria (supporting evidence) that each source database uses, with each update, to avoid unexpected expansion or contraction of markers included in the platform.
3.2. Tools for antimicrobial resistance prediction from sequence data
Potential AMR detection tools that the PATH-SAFE platform could draw on are 1) AMRFinderPlus utilising the NCBI RefGenes database (National Institute of Health (US), 2024a), 2) ResFinder drawing on the ResFinder database (DTU Food, 2022), 3) CARD RGI using the CARD database (Alcock et al., 2023), 4) abritAMR which runs AMRFinderPlus and processes the results with a Python wrapper script (Horan et al., 2023), and 5) Pathogenwatch which has the internal functionality to screen against a dictionary of AMR determinants and mapping them to drugs and drug classes (Centre for Genomic Pathogen Surveillance, 2018). Features of abritAMR and ISO-certification will be discussed in Section 5 on international best practices and data standards for AMR surveillance in FBD.
ResFinder has functionality for raw read analysis which is useful to detect azithromycin resistance mutations on the 16S ribosomal RNA in Salmonella (Nair et al., 2016). ResFinder utilises the novel ConClave algorithm as part of the KMA tool which was specifically developed for this task. It applies an additional mapping step that identifies template candidates and lowers the computational requirements for the AMR analysis of raw sequence data (Florensa et al., 2022).
ResFinder and AMRFinderPlus can be considered critical infrastructure for AMR surveillance. Such critical infrastructure ought to be sustained through official dedicated funding streams in light of their utility to national and international agencies. Although required for EU surveillance programs, ResFinder is currently dependent on in-kind funding from its host institution (Technical University of Denmark, DTU) or funding from donors which affects sustainability. AMRFinderPlus and its associated databases are part of the NCBI Pathogen Detection program (see 5.2.1), funded by the US government as core infrastructure to support national agencies (including NIAID, CDC, FDA, FAO) in addressing the National Action Plan for AMR, although it is relied on by a global user base including national-level agencies in many other countries. Long-term funding is critical to sustain stable platforms, databases and tools for transparent and reproducible analysis of surveillance data. This is an important consideration for future sustainability of PATH-SAFE as a national public health program for the UK.
Recommendations:
-
For simplicity of software engineering, the platform should implement existing tools rather than creating another marker database and drug dictionary that need to be tested and maintained.
-
For interoperability with international agencies, the platform should facilitate comparability with both ResFinder and AMRFinderPlus output, for the reasons outlined above.
-
Use of abritAMR as a wrapper for AMRFinderPlus is not currently recommended for the platform as
-
it is complex to modify abritAMR reporting logic for local needs; and
-
its use creates a dependency on abritAMR to maintain updates to the AMRFinderPlus software, before such updates can be reflected in the platform.
- Sustainable databases and analysis platforms for interpretation of WGS surveillance data need long-term reliable funding, reflecting their status as essential infrastructure for national public health agencies.
Recommendations on reporting:
-
The platform should report confident AMR markers organised in the form of an antibiogram, i.e. grouped by antimicrobial rather than drug class as far as possible, in order to be comparable to phenotype data.
-
ResFinder implements an internal dictionary mapping determinants to drugs, making it the preferred tool. AMRFinderPlus includes mapping to drug class but currently lacks a coherent mapping to drug level.
-
Nevertheless, given the widespread use of AMRFinderPlus, consideration should be given to how results from this tool can be combined or compared with those of ResFinder, within the platform.
-
The platform documentation should make clear with which parameter values each tool is run and provide versioning with each output.
-
The platform should make accessible to users the full summary results of AMR marker searches (including % coverage and identity of AMR gene hits, and accession of the query gene sequence) as an optional file download, so that users may optionally investigate confidence in the resistance marker calls underlying the summary antibiogram. Standardisation of platform outputs should be considered, especially if multiple tools (ResFinder and AMRFinderPlus) are implemented, to facilitate straightforward downstream use.
3.3. Validation of antimicrobial resistance prediction tools
Validation of genomic AMR prediction tools is essential for their use in FBD surveillance. There are three ways to conduct a validation, 1) using an alternative genomic analysis method than those used by the prediction tools, 2) conduct PCR analysis for key resistance markers, or 3) compare genotypic resistance predictions to phenotypic resistance profiles.
Recommendations:
-
Validation should be done for isolates from all sectors involved in food security, as markers may be distributed differently.
-
Comparison to phenotypes is not within the scope of this platform which is geared towards cross-sector surveillance. It is envisaged that agencies will conduct phenotyping in addition to sequencing where this is considered important. Validation could be done by stakeholders against their own current phenotyping methods, if considered important for them.
-
Comparison to PCR detection is relevant only insofar as PCR is currently used by stakeholder agencies, and in such cases comparison to PCR as current practice would be important for ISO accreditation. As PCR is not used for AMR marker detection by UK agencies currently, it is not a priority to validate in this way.
-
Validation against alternative genomic analysis methods should focus on comparison to ResFinder and AMRFinderPlus. If the platform implements ResFinder, it should be validated against AMRFinderPlus, and any differences documented so that users are aware. If both tools were to be implemented in the platform, this validation would essentially be in-built.
-
Comparison to alternative genomic analysis methods should focus on comparison to outputs of existing pipelines currently utilised within the stakeholder agencies, so that differences can be understood and potentially addressed (as these approaches are likely to be run in parallel there is potential for confusion).
4. Data standards for genomic surveillance of foodborne diseases
Data standards are critical to enable comparability and meaningful joint analysis of different datasets on a shared platform. Two key issues that need to be addressed to implement WGS surveillance of FBD are harmonised metadata requirements and the tension between collecting and reporting enough metadata to conduct meaningful investigations and privacy concerns around the identifiability of individuals, farms or commercial entities. Moreover, QC metrics for sequence data and reporting criteria need to be defined. Metadata, QC metrics and reporting criteria should meet the needs of the end user group and if possible allow for comparability with data produced by international agencies, while the data privacy framework needs to take into consideration local regulations. This section makes recommendations on metadata and QC requirements, reporting criteria and user access policies for the platform, assuming that data will primarily be shared among public health and government agencies. Data sharing by the food industry is not precluded, but it has not been considered as part of these recommendations and should be part of a separate discussion held as part of the Food Safety Research Network initiative.
4.1. Metadata requirements
Institutions involved in FBD and AMR surveillance collect a wealth of data but cannot share all their data indiscriminately with other entities. Institutions use varying metadata ontologies that are currently not harmonised. Since the PATH-SAFE platform is meant to facilitate data sharing among institutions, a minimum unified metadata set to be submitted to the platform together with sequence data needs to be defined. The scope of the metadata collected by the current PATH-SAFE platform is intentionally narrow to ensure the minimum data is collected, stored and processed in line with UK GDPR guidelines. At the same time, this will facilitate communication and data sharing between agencies. The platform will be expansible so that new data fields can be added later.
The minimum metadata requirements should be compatible with the metadata collected by each agency and are not meant to replace them. Moreover, the minimum metadata set should allow one agency to contact another agency, for example, if during an outbreak investigation it is revealed that the genome sequences submitted by the two agencies fall into the same cluster. In this scenario, it will be important that the minimum metadataset has variable fields for linking sample, sequence and subject IDs between submitting agencies. Appendix 2 includes a table with the proposed minimum metadata requirements. The minimum metadata should align with the international standard ISO 23418:2022 on metadata requirements for WGS for typing and characterisation of bacteria (International Organization for Standardization, 2022b). In addition, an extended metadata set with a restricted access policy for authorised users only may be defined.
To facilitate metadata reporting, templates for different types of specimens can be designed and standard ontologies of terms to be completed for each metadata field should be used. For example, the US cross-agency Genomics for Food Safety (GenFS) Metadata Workgroup has recently published a One Health metadata template (National Institute of Health (US), 2024d; Pennerman & Timme, 2024). Reporting whether AMR specimens are derived from the human, animal or environmental sector can help to track how AMR pathogens are transmitted between sectors (Feldgarden et al., 2021). The UKHSA has recently published a framework for genomic pathogen surveillance, including One Health, that the PATH-SAFE platform should build on (UK Health Security Agency, 2024).
To prevent identifiability of personal or commercially sensitive information, it is possible that not all fields in the minimum metadata set need to be completed. In this case, it will be important to have a defined priority of which fields need to be completed and which can be omitted to protect sensitive information. The platform needs to be flexible enough to accommodate changing metadata requirements and potential expansion of metadata fields in the future.
Recommendations:
-
The number of metadata fields should be small initially to facilitate upload of data and use of the PATH-SAFE platform and to be consistent with UK GDPR obligations.
-
Minimum metadata requirements of the platform should be compatible with the metadata collected by each agency. They should also be compatible with concerns around data sharing and legal obligations.
-
The platform is expected to be expanded at a later date. Therefore, it should be built in a way that allows new metadata fields to be incorporated later as the need arises.
-
Where possible, metadata fields should use standardised ontology terms.
-
Metadata should have variable fields for linking sample IDs with submitting agencies to allow one agency to contact another to request access to relevant data. The PATH-SAFE platform could provide a secure environment for organisations to share data with trusted partners.
-
The platform should provide a mechanism for the generation of anonymised sample IDs and provide a simple mechanism for the publication and sharing of sequence data with public resources in a way that is compliant with UK GDPR.
-
All metadata must be processed in line with UK GDPR guidelines and align with organisational policies and relevant legislation.
4.2. Quality control requirements
All genomes assembled by the PATH-SAFE platform’s assembly pipeline will be added to the database but hidden if they fail read or assembly QC thresholds. Table 3 lists proposed QC thresholds for inclusion of genome assemblies. Prior to assembly, genomes will be filtered out if they have less than 50x coverage. Thresholds for genome size and GC content have been derived from an analysis of the 1239 complete Salmonella enterica genomes in the NCBI Reference Sequence (RefSeq) database (National Institute of Health (US), 2024c). Analysis of the 12,698 whole genome sequences in the NCBI Reference Sequence database arrives at a similar but somewhat wider range of values. The proposed threshold values for other QC metrics are commonly used in WGS surveillance (e.g. 44).
The platform will have an in-built mechanism validating each metadata field to prevent errors in the submitted data. Each field will have a range or set of permitted values. For example, the field for reporting date of sample collection will only accept values in a standardised date format, such as 2023-05. Values that do not conform to this format or are outside of a realistic range, for example, 1023-05, will be flagged. Permitted values may be expanded over time if necessary. To prevent identifiability, the platform will apply the principles of data minimisation and obfuscation. For dates, instead of the precise date of sample collection, only month and year may be displayed if required. If an error occurs during data upload the platform should offer functionality to update the incorrect data field without having to reupload the entire dataset.
Recommendations:
The PATH-SAFE platform should have an automated QC mechanism and the criteria in Table 3 should be used by the platform for validating data.
-
The PATH-SAFE platform should have an automated QC mechanism and the criteria in Table 3 should be used by the platform for validating data.
-
There should be a mechanism built into the platform to validate metadata uploaded.
-
The platform should offer the option to keep variable values fuzzy to prevent identifiability of individual samples.
-
For ease of use, options for both bulk upload of metadata and for upload of individual metadata fields should be offered by the platform.
-
The platform should include options to update individual metadata fields in case they contain incorrect values.
4.3. Reporting criteria
Reporting criteria should be defined by organism of interest and use case. The defined reporting criteria should be included in the output generated by the platform as well as the date on which databases were queried and the methods used.
Recommendations:
-
For the sake of transparency, species- and case-specific reporting criteria for WGS analysis of FBD should be developed by subject matter experts.
-
Reporting criteria should be in alignment with those of agencies involved in FBD surveillance in the UK.
-
Reporting criteria should be included in the output generated by the platform.
4.4. Access policies and data sharing among agencies
To ensure the security of, and to build trust into the PATH-SAFE platform, transparent data access policies need to be implemented and data access by users needs to be audited. User access policies for a similar platform, CLIMB-TRE, have been adapted from those developed by the COG-UK consortium during the COVID-19 pandemic. These access policies have been agreed on previously by UKHSA and other public health agencies for similar data (genomic data and metadata similar to those chosen for PATH-SAFE). These user access policies are still in use and can serve as a template for the PATH-SAFE platform.
CLIMB-TRE user access policies account for four different user types; public health agencies and service users, government agencies, researchers, and submitting laboratories. The latter are laboratories that are commissioned to sequence COVID-19 samples and that have permission to upload data to the CLIMB-TRE platform but have no access to output or other parts of the system. Different access levels for different users or user types allow for granular access control. There are two principal options for implementing different user access levels:
-
create default levels of sharing designed to allow agencies to access data without requesting it,
-
all agencies have access to their own data and the option to grant granular access to other users.
Once metadata requirements and user access levels have been defined, agencies need to agree on access levels for different users. Where access priorities differ among agencies, the highest priority level required by any one agency should be implemented.
On the platform all submitted samples will have a unique anonymous identifier. The platform will by default, run analysis on all data that a user has access to. This is relevant, for example, if during an outbreak samples are submitted by different agencies.
Access levels should also conform with UK GDPR requirements which will ensure safe management of sensitive data, as well as commercially sensitive data for FBD surveillance. Moreover, access can be granted in a stepwise manner. For example, in a first step users can be granted initial access to the platform and in a second step agencies can collaborate with further data sharing needs for each investigation. Access policies will be decided by a group that will contain representatives of all participating public health agencies.
The development of the user access policies needs to consider the Five Safes principles (Office for National Statistics, 2017). These include:
-
Safe people: new users seeking access to the platform need to demonstrate that they have the technical skills for the access level they request and agree to abide by the data protection rules of the platform and participating agencies.
-
Safe projects: users requesting access to data have to submit a proposal outlining how data will be used in an ethical and appropriate way to answer research questions that will generate public benefits.
-
Safe settings: users do not receive a copy of the data. Instead, all analysis needs to take place in the secure environment of the PATH-SAFE platform.
-
Safe outputs: output generated from data on the platform needs to conform with the same data protection rules as the original data. This needs to be checked prior to exporting outputs from the platform.
-
Safe data: data used for analysis on the platform need to be anonymised or pseudonymised, if anonymisation is not possible.
A common framework agreement for data sharing among UK agencies involved in public health and environmental health already exists, including for biosecurity under the Four Nations Health Protection Oversight Group (HPOG). This framework addresses legal issues involved in data sharing among UK public health and government agencies. However, there is a need for standardised data sharing templates for different purposes and user access levels to facilitate and accelerate data sharing among different users of the PATH-SAFE platform going forward.
Recommendations:
-
The PATH-SAFE platform needs transparent user access policies and functionality to audit who has accessed what data.
-
Different access levels for different user types should be implemented.
-
The development of user access policies should be based on the Five Safes: safe people, safe projects, safe settings, safe outputs, safe data.
-
The platform should implement standardised data sharing templates for different user access levels to facilitate data sharing among different UK agencies involved in genomic surveillance of FBD and AMR.
5. International best practices and reporting standards in genomic surveillance of foodborne diseases
As FBDs do not stop at borders, international coordination and sharing of WGS data will be critical to understand and curb the global spread of foodborne pathogens. Strategies for molecular surveillance of FBD have been developed and implemented by public health agencies in multiple countries and are supported by the WHO, FAO and other international agencies and initiatives (Food and Agriculture Organization of the United Nations, 2024; World Health Organization, 2023). Countries are at different stages of implementing molecular surveillance for FBD and differ in their regulations on when sequencing of isolates is required and what metadata are reported. FBD and AMR surveillance in Europe is coordinated by ECDC and EFSA (see Section 5.1.3) (World Health Organization, 2023). To facilitate sharing of WGS surveillance data with global stakeholders and partners, data standards have been developed by the Genomic Standards Consortium (GSC) and the Public Health Alliance for Genomic Epidemiology (PHA4GE).
To facilitate the timely detection of FBD outbreaks at the international level, PulseNet International was formed, an international network of laboratories working in the food- and waterborne disease and One Health sectors to improve the investigation of food- and waterborne disease outbreaks and food safety (see Section 5.1.1 on how PulseNet is integrated in the genomic FBD surveillance workflow in the US) (Nadon et al., 2017). Institutions participating in the network can upload WGS data associated with FBDs to the network’s online database which allows for the identification of clusters of FBD-associated isolates from different sectors in the food supply chain across international borders.
The International Organization for Standardization is another important player in the genomic FBD surveillance space, as it develops standards for quality control and quality assurance of tools, standard operating procedures (SOPs) and workflow used for genomic surveillance in the food chain and in clinical and public health microbiology laboratories (see Section 5.1.2) (International Organization for Standardization, 2024).
This section provides an overview of existing national and international strategies for molecular surveillance of FBD and presents examples of best practices for molecular FBD surveillance in different countries.
5.1. Best practices for genomic foodborne disease and antimicrobial resistance surveillance - international examples
This section gives three international case studies meant to illustrate best practices for genomic surveillance of FBDs and AMR. Key lessons learnt are summarised after each case study.
5.1.1. Implementation of genomic epidemiology and antimicrobial resistance surveillance in foodborne diseases by the US Food and Drug Administration
Three agencies in the US are involved in food safety surveillance: the Food and Drug Administration (FDA), the Department of Agriculture Food Safety Inspection Service (USDA-FSIS), and the Centers for Disease Control and Prevention (CDC) (Stevens et al., 2022). In 2013, the FDA initiated the GenomeTrakr network that today includes approximately 50 national and international laboratories, all sharing WGS data via the National Center for Biotechnology Information Pathogen Detection (NCBI-PD) database (Allard et al., 2016; Timme et al., 2020). The NCBI database is a long-standing fully funded piece of data infrastructure used by non-US entities, including PHE/UKHSA, for data upload and download. NCBI, along with the European Nucleotide Archive (ENA) and the DNA Databank of Japan (DDBJ) comprise the International Nucleotide Sequence Database Collaboration (INSDC) (Cochrane et al., 2011).
The GenomeTrakr Program maintains a suite of open source resources, adhering to the FAIR (Findable, Accessible, Interoperable, Reusable) data standard. GalaxyTrakr, an open access analysis platform, provides users a variety of custom genomic epidemiology tools, including quality control workflows. GenomeTrakr also maintains a public, version-controlled workspace for protocols (protocols.io) that provides detailed steps for the entire whole genome sequencing workflow.
The FDA’s genomic epidemiology workflow comprises multiple agency arms and links to the US CDC, USDA-FSIS, states health and agriculture departments, and the network of submitting laboratories (Figure 2). Within the FDA, the Office of Regulatory Affairs is responsible for inspections and sequencing, the GenomeTrakr Program within the Office of Regulatory Science is responsible for overseeing data collection and NCBI submission across the laboratory network, the Office of Analytics and Outreach watches for signals in the submitted data and is responsible for data interpretation and risk assessment. If a signal is detected the Office of Compliance and the Coordinated Outbreak Response and Evaluation (CORE) Network are alerted and initiate communication with the CDC, USDA and relevant state departments. CORE and the Office of Compliance may draw on the Office of Regulatory Affairs to undertake inspections or product testing. This workflow is supported by the Office of Food Safety which is responsible for policy development, and the Office of International Engagement (OIE) which is responsible for communications with food security and public health related agencies in other countries.
To improve data sharing among agencies involved in food security surveillance in the US and to make surveillance and outbreak investigations more effective and efficient, the FDA and other collaborating national and international partners developed standard data structures for submitting and storing pathogen data at the INSDC. These include the Pathogen Data Object Model, the One Health Enteric metadata package, and the FoodON ontology for terms submitted to the metadata templates (Dooley et al., 2018; Pennerman & Timme, 2024; Timme et al., 2023).
An economic evaluation of the GenomeTrakr programme estimated that by 2019 the net monetised annual health benefits derived from the programme exceeded US$ 500 million, compared to an investment of US$ 22 million by public health agencies. This corresponds to a return on investment of US$10 in averted human health costs for every US$1 invested (Brown et al., 2021). This finding is in line with an economic evaluation of WGS for pathogen surveillance by UK and EU institutions that also demonstrated a net benefit of the additional information gained from genomic analysis, although this study found considerable variation in the cost of sequencing technologies across institutions (Alleweldt et al., 2021).
Key lessons learnt:
-
Open access platforms and open source protocols facilitate data sharing nationally and internationally.
-
A standardised regulatory workflow turns insights gained from genomic epidemiology into impactful food safety/public health interventions.
-
Structured and harmonised metadata ontologies improve prediction accuracy and make interagency collaboration more efficient, decreasing time to response.
-
When implemented effectively, genomic epidemiology of FBD pathogens is cost-effective with a high return on investment of 10:1.
5.1.2. abritAMR, an ISO-certified genomics workflow for antimicrobial resistance identification and surveillance
There is currently a lack of international standards for bioinformatic tools and pipelines for detecting AMR mechanisms. The International Organization for Standardization (ISO) supports the harmonisation of processes and validation for quality management. Most ISO standards that affect AMR identification are aimed at wet lab work (International Organization for Standardization, 2022a), although recently the first standard for WGS data generation and analysis from bacteria from the food chain has been published (International Organization for Standardization, 2022b) (ISO 23418 : 2022). A second ISO for AMR analysis in food products is being launched.
Standardisation and accreditation generate trust and acceptance of novel analytical tools and pipelines and assure end users that they obtain actionable information (Ballard et al., 2023). Therefore, many governments require standardisation and formal accreditation of clinical and public health laboratories. For example, the EU expects all National Reference Laboratories to accredit all methodologies and processes used, including WGS.
5.1.2.1. Details of the workflow and validation
In Australia, the Microbiological Diagnostic Unit Public Health Laboratory and the Department of Health Victoria have developed, validated and implemented abritAMR, a bioinformatic tool that provides a wrapper for the NCBI AMRFinderPlus tool for the detection of AMR determinants from WGS data (Sherry et al., 2023). The wrapper adds a classification step and reporting logic to produce tailored reports on AMR determinants of clinical or public health importance. abritAMR consists of five components:
-
NCBI’s AMRFinderPlus tool to identify AMR determinants in sequences,
-
NCBI’s ReFgenes database, a curated AMR determinant sequence database,
-
A classification database to translate between AMRFinderPlus output and relevant public health and clinical reporting criteria,
-
A species-specific reporting logic for AMR determinants,
-
An inference mechanism for AMR from sequence data (currently only validated for Salmonella)
ISO-certification was undertaken for the abritAMR workflow, SOPs and pipeline code to demonstrate adherence to standards in clinical and public health microbiology laboratories. Validation was carried out using isolates obtained from routine AMR surveillance in public health laboratories using predefined thresholds for acceptable sensitivity and accuracy. Three validation steps were undertaken, comparing the abritAMR output to 1) PCR results for AMR genes, 2) Sanger sequencing for allelic variants, 3) synthetic read sets generated from short-read sequence data from complete, publicly available genomes from RefSeq or GenBank. The synthetic dataset is also used as the “abritAMR test suite” for re-verification (see below). In these validation tests, abritAMR showed very high accuracy of 99%, with 97.9% sensitivity and 100% specificity. False negatives were mainly due to AMR genes being fragmented across multiple contigs resulting in partial matches or to loss of AMR plasmids in culture (Sherry et al., 2023).
Inferred antibiograms were validated using a set of 864 Salmonella isolates that had both AST data and WGS data. In this test, abritAMR showed 98.9% accuracy, 98.9% sensitivity and 98.9% specificity. Discrepancies were due to the detection of partial genes at contig breaks or difficulties with resolving sequences with multiple alleles of the same gene family which may be collapsed into a single gene detection by abritAMR or miscalled as a different allele. Use of alternative assemblers had a minimal impact on validation results but should be considered in the case of discrepancies (Sherry et al., 2023).
Precision testing was carried out on a test panel of 13 organisms using different sequencing platforms, a range of sequencing throughput modes and read-lengths. The limit of detection, defined as the minimum average coverage required for accurate detection of gene targets or alleles, was determined by varying the coverage from 40x (minimum required for QC purposes) to 150x. Even with a coverage of 40x high accuracy for AMR detection could be achieved (Sherry et al., 2023).
According to ISO standards, re-verification is required after every update of the AMR database or AMR detection tool. A re-verification with the abritAMR test suite is undertaken after every minor update that does not affect the core logic of the tool or workflow. A full re-verification with all steps described above is carried out after major updates that change the core logic of the workflow or the structure of the output (Sherry et al., 2023).
After validation of the abritAMR workflow, it was implemented in routine surveillance activities by modifying reporting outputs, integration with existing Laboratory Information Management Systems, and documentation of all standard operating procedures. Consultations were held on the proposed workflow output, including with domain experts to ensure that results are interpretable and meet reporting obligations for different pathogens, quality management staff to ensure that workflow outputs meet legal requirements, and end users to ensure that outputs deliver actionable insights (Sherry et al., 2023).
The outcome of building and validating the abritAMR tool is a routine, streamlined and rapid AMR detection workflow. A sequencing run with 96 samples takes just three minutes on 256 CPUs. Moreover, the new validated workflow reduced the need for manual reclassification of fields that should be included in reports and those that should not, for example, by removing intrinsic AMR genes from reports (Sherry et al., 2023).
Key lessons learnt:
-
Validation is necessary to generate acceptance and trust in WGS surveillance, and accreditation is required in some countries.
-
There is a lack of international standards for bioinformatic tools for WGS surveillance, but new standards are in preparation, for example, by PHA4GE.
-
Accreditation of AMR genomics workflows can improve comparability of the standard of AMR reporting across countries.
-
Validation and benchmarking of WG databases, tools, and pipelines for AMR detection requires a broad range of publicly available reference datasets with genotypic and phenotypic data.
5.1.3. Implementing genomic epidemiology in the EU - the example of Denmark
Denmark has implemented genomic pathogen surveillance since 2010 when the Center for Genomic Epidemiology received funding for initially six years. From 2016 onwards Denmark has implemented routine genome sequencing of pathogens following ECDC and EFSA guidelines. The three institutions involved in genomic epidemiology in Denmark are the Statens Serum Institut (SSI), the Danish Veterinary and Food Administration (DVFA) and the Technical University of Denmark (DTU), managed by the Central Outbreak Management Group (DTU National Food Institute, n.d.).
The data management structure for zoonotic disease surveillance was restructured three years ago to streamline data sharing and collaboration among researchers, healthcare professionals and industry. Institutions that generate data also host it and have the option to share their data for joint analysis within the outbreak surveillance group. The underlying principle of the new data management and analysis infrastructure is to bring the data to the tools. The joint space for data sharing and analysis is the Computerome 2, one of Scandinavia’s largest HPC (high performance computing) clusters, which offers a large storage and compute capacity and access to standardised bioinformatic analysis tools (https://www.computerome.dk/). Organisations can upload their data into this joint space and use the tools and compute capacity to analyse them. Other public health institutions also use the Computerome 2, and each organisation has its own space on the HPC cluster. Institutions typically share sequence data in the joint space, while potentially sensitive metadata continue to be hosted by the data holding institution.
Sampling plans for FBD organisms and AMR analysis in Denmark follow EU regulations and are implemented under the Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP, Figure 3). Surveys are conducted in animals at slaughter and meat and there are recommendations on the minimum number of isolates to be analysed. All Salmonella samples from humans, animals and food sources are sequenced and analysed for AMR markers. WGS is also performed on Campylobacter, Listeria and ESBL, ampC and carbapenemase-producing E. coli. Neither ECDC nor EFSA require collection of genomic data for AMR surveillance. According to EU regulations for One Health surveillance, only ESBL/ampC-producing E. coli isolates recovered from livestock animals and meat and from humans with bloodstream infections need to be genotyped to map the frequency of MLST and AMR genes.
In addition to routine surveillance in animals and food, investigative surveys for AMR in specific foods or populations may be performed to understand the risk to consumers from new sources, to look for new or changed resistance profiles and to inform discussions among policy makers, scientists and industry. Supplementary sampling may be conducted to fulfil EU requirements or to fill gaps in EU surveys in traditional populations. Moreover, data from control programmes, clinical diagnostics or research programmes also serve as input to surveillance reports.
FBD and AMR surveillance in humans is informed by samples collected in general practices and hospitals and examined by the Department of Clinical Microbiology and reference laboratories at SSI. Data from these samples are collated at SSI through laboratory information management systems (LIMS) and the Danske Mikrobiologi Database (MiBa). As data from the veterinary and food sector and from the human sector are collected by different national agencies and reported to different supra-national agencies, harmonisation of databases is important to enable interoperability between different components of the surveillance system.
Key lessons learnt:
-
To collaborate effectively, different agencies involved in FBD surveillance need a joint analysis space where they can “bring their data to the tools”.
-
This analysis space needs to offer a highly secure and controlled environment where agencies can share WGS data while maintaining control of potentially sensitive metadata.
-
The development of standardised open access tools supports national and international surveillance efforts and collaboration.
-
Harmonisation of sampling plans and data formats between national and supra-national agencies is necessary to ensure interoperability of data systems.
5.2. Public and international data sharing
Sharing of WGS data among agencies that will use the platform (see Section 4.4), but also with international collaborators, has important and measurable benefits in terms of identifying outbreak clusters and the extent of their spread (see Section 5.2). Publicly available genomic databases are also important to develop and validate new bioinformatic tools and to place new genome data into a historic context.
There is still considerable concern around data sharing in public health and regulatory agencies in many countries. The main worry is that improper use of data may lead to economic or reputational damage, especially for data from animal and crop isolates linked to industry. The frequently cited example in this context is the 2011 E. coli O104:H4 outbreak in several European countries that was first incorrectly attributed to cucumbers originating from Spain but was later confirmed to be attributable to sprouts from Egypt (Buchholz et al., 2011; Frank et al., 2011; King et al., 2012). Import bans on Spanish vegetables cost the country an estimated £120 million per week (British Broadcasting Corporation, 2011). However, this is the only documented incident of this nature, and analysis protocols and tools have improved significantly since 2011. Generally, it is fair to assume that anonymised data will be used responsibly by competent experts in public health agencies and subject to risk assessment and hypothesis generation, including for cross-border investigations into widely distributed food products.
A positive example illustrating the benefits of WGS data sharing is the 2022 Salmonella enterica outbreak in Kinder Surprise products (Larkin et al., 2022) In this event, the UK identified a single-linkage cluster characterised by an unusual AMR pattern that had not been observed in human disease, livestock or food in the UK before. Following the UK’s notification on ECDC’s EpiPulse Food and Waterborne Diseases platform, Germany, Sweden, France, the Netherlands and subsequently Luxembourg, Norway, Ireland, Belgium and Spain reported confirmed or probable cases. The European Union agreed on a case definition based on genomic information accounting for differences in genomic methodology in member countries. Food chain investigations could link the outbreak to a single production site in Belgium, where the outbreak strain had been detected in processing equipment in December 2021, so that outbreak control measures across multiple countries could be implemented. The outbreak investigation and response demonstrate the benefits of sharing microbiological sequence information from multiple sectors (human disease, environmental sampling) and cross-country collaboration.
WGS data is distinct from other types of public health data in that it is digital by nature and has broad utility for international public health agencies, R&D by industry, and academic research by a broad range of disciplines. Thus, the bar for sharing WGS data is comparatively low, and WGS data generated by public funds can be expected to be made publicly available in a timely fashion. For example, GenomeTrakr requires sequence data generated by projects supported by US Government funding to be uploaded to the public NCBI database and encourages institutions to submit WGS data as soon as the sequencing runs have finished (National Institute of Health (US), 2024b). To facilitate submission of metadata, GenomeTrakr provides contextual metadata templates for clinical or host associated data, environmental or food associated data, combined submissions and a One Health template for enteric pathogens (National Institute of Health (US), 2024b). GenomeTrakr has existed for more than a decade and has not experienced any incident where data was used inappropriately in a way that led to harm to the originator of that data.
UK agencies involved in FBD surveillance are supportive of WGS data sharing. For example, UKHSA (formerly Public Health England) maintains a NCBI BioProject with over 65,000 Salmonella genome uploads released to the public domain (https://www.ncbi.nlm.nih.gov/bioproject/248792). This ongoing practice should be continued and would ideally be improved on by reducing time to data release. Currently, data may in some instances be released years after the outbreak in which they have been collected.
Recommendations:
-
Data sharing among national agencies and international collaborators has important benefits for identifying outbreak clusters and the extent of their spread, to develop and validate new bioinformatic tools and to place new genome data into a historic context.
-
Although there is considerable concern about potential negative consequences of sharing of WGS data of foodborne pathogens, especially with respect to industry, there is no evidence of these negative consequences materialising.
-
WGS data generated with the support of public funds should be made publicly available, if possible, in a timely manner to allow real-time analysis of ongoing outbreaks, and if possible, with sufficient contextual metadata to be useful for public health.
6. Conclusion
The application of genomics in FBD surveillance has increased substantially over the past years, especially for high-resolution pathogen typing and cross-sectoral outbreak investigations but also for AMR prediction, especially for Salmonella pathogens (National Institute of Health (US), 2024b). Consequently, WGS surveillance is expected to increasingly inform interventions for food safety and public and veterinary health. Addressing the threat from FBD and AMR will require cross-sectoral interventions and data sharing between agencies and laboratories at the local, regional and national level. Harmonisation of WGS data standards and analysis methods among agencies involved in FBD surveillance is crucial to facilitate data sharing, interpretation of results and accelerate the implementation of effective and targeted interventions.
The PATH-SAFE platform will provide a secure and controlled environment for UK agencies involved in FBD surveillance to share WGS data, use standardised analysis tools and collaborate. The platform is not meant to replace but to complement existing agency-internal solutions and should easily integrate with existing workflows.
This technical note summarises recommendations made by Community Input Advisory Groups on the design and capabilities of the platform and on the application, technical requirements of and best practices for WGS in FBD surveillance in general.
Documentation of the PATH-SAFE platform and validation reports will be provided in separate documents.
7. Biographies of Community Input Advisory Group Members
Dr. Marc W. Allard is a Senior Biomedical Research and Biomedical Product Assessment Services Officer in the Division of Microbiology in FDA’s Office of Regulatory Science. Dr. Allard joined The US FDA in 2008 where he uses Whole Genome Sequencing (WGS) of foodborne pathogens to identify and characterise outbreaks of bacterial strains, particularly Salmonella, E. coli, and Listeria. Dr. Allard specialises in both phylogenetic analyses, as well as the biochemical laboratory methods which generate the WGS information. Dr. Allard helped develop the first distributed network of laboratories that utilize whole genome sequencing for pathogen identification and traceback called the GenomeTrakr database, which is part of the NCBI Pathogen Detection web site. These tools are used daily for outbreak investigations and compliance. Dr. Allard acts as senior scientist to advise the US FDA on both WGS and phylogenetic methods as they apply to public health.
Dr. Marie Anne Chattaway, a seasoned Clinical Scientist, serves as the Head of the Salmonella reference service at the Gastrointestinal Bacteria Reference Unit (GBRU) within the UK Health Security Agency, boasting an extensive tenure of over two decades. Beyond her main role, she actively engages in surveillance initiatives, providing vital microbiological information and expertise for outbreak investigations, and conducts in-depth analysis of genomic data, particularly focusing on antimicrobial resistance and population structures. Currently, she holds the position of co-lead and career academic lead for the Genomic and Enabling Data Health Protection Research Unit (GED HPRU). Dr. Chattaway’s commitment to global health is evident through her decade-long involvement in fortifying laboratory capabilities for enteric bacterial diseases across Africa, achieved through the development and implementation of comprehensive training programs, all in support of the International Health Regulation program’s Global Health initiative. Additionally, she is currently undergoing training to become a consultant microbiologist, further solidifying her expertise and impact in the field.
Professor Tom Connor is the Head of the Public Health Genomics Programme within Public Health Wales, the national Public Health Institute of Wales, and is a Professor of Bioinformatics and Pathogen Genomics at Cardiff University. He earned his PhD from Imperial College, before undertaking a PostDoc at the Wellcome Sanger Institute. Following this, in 2012, he took up his permanent academic position at Cardiff University. He has a research track record that has seen his team apply genomics to examine microbial populations, to understand their evolution and, in the case of pathogens, their genomic epidemiology. As an extension to his research work, Tom has provided leadership as part of the development of national computational infrastructures - such as the MRC CLoud Infrastructure for Microbial Bioinformatics (CLIMB) and Supercomputing Wales - which are designed to support genomics and bioinformatics research. Since 2016 Tom has worked to translate his research expertise into clinical service, as part of his role within Public Health Wales. Within Public Health Wales, Tom has led the development of bioinformatics as an area of activity, as well as developing the pipelines and infrastructure that underpin current clinical services. As part of this work Tom has overseen the development of ISO 15189 accredited clinical genomics services including for C. difficile (first service of its type in the world), HIV (first national service of its type in the UK), SARS-CoV-2 (first accredited SARS-CoV-2 genomics service in a UK public health agency). Collectively these services have sequenced almost 300,000 pathogen samples from Welsh patients to support services ranging from individual patient management to national-scale pandemic monitoring and response.
Dr Tim Dallman is an international expert in pathogen genomics, phylogenetics and bioinformatics with respect to foodborne pathogens. He is currently an Assistant Professor at the Institute of Risk Assessment Sciences at the University of Utrecht where his research interests focus on genomic analysis of foodborne pathogens and their environment from a one health perspective. Previously he worked for over ten years at UKHSA as the Lead for Bioinformatics of Gastrointestinal Pathogens and led the implementation of whole genome sequencing methodologies into reference microbiology and infectious disease surveillance He is a regular expert participant at working groups on food safety and genomics organised by international organisations including ECDC, WHO and UN-FAO and has had considerable interactions with the food industry to help inform how genomic microbiology can be leveraged in this sector and how it can feed into risk and assessment and public health surveillance.
Professor Alistair Darby is Co-Director of the Centre for Genomic Research (CGR) at Liverpool and has more than 15 years experience as an applied entomologist. His main area of research is in microbial/host interactions which covers his interest in arthropod symbiosis, pathogens and microbiomes. He has extensive experience in designing and analysing genomic and RNAseq data, including arthropod genomes; e.g., Nasonia wasp (Science), tsetse fly (Science), mite (GigaScience), and a new high quality version of the Diamondback moth genome sequence using the PacBio (data at lepbase.org). His research also uses single cell genomics and he is the lead for the CGR single cell genomics facility.
Dr Tigan Daspan is Head of Digital Transformation at Food Standards Scotland. His involvement in the DAG is mainly from a data protection, information security and data quality & governance perspective.
Dr Richard Ellis is Head of Genome Analysis at the Animal and Plant Health Agency. After completing his DPhil from Oxford, he spent over 10 years in academia studying bacterial ecology and population genetics before joining APHA. His work now focuses on the application of high-throughput sequencing technologies for understanding the evolution and spread of pathogens, for investigating diseases of unknown aetiology, and the integration into disease surveillance programmes. He oversees the laboratory teams responsible for routine disease testing required for both surveillance and trade, and the capacity to scale up testing in the event of a disease outbreak.
Professor David Gally holds a personal chair in Microbial Genetics at the University of Edinburgh and has been part of the Roslin Institute since 2011. His background is in microbiology and food safety. He was seconded to Food Standards Scotland as their Chief Scientific Advisor from 2021-24. David has led three successive Roslin Institute Strategic Programme grants funded by the BBSRC, the latest on the ‘Control of Infectious Diseases’ in Livestock (2017-2022). His group has been applying machine learning to source attribution for bacterial isolates associated with human infections through food or water, in particular Salmonella and E. coli, to help with outbreak investigations and ongoing work has moved into predictive phage therapy.
Dr Matthew Gilmour leads the ‘Listeria and other Invasive Pathogens’ research group at the Quadram Institute in Norwich, England. Matthew is also deputy lead of Quadram’s ‘Microbes and Food Safety’ strategic programme which has a focus on translating the Institute’s key microbiology findings and genomic technologies with partners in the food industry and government.
Matthew was previously based in Canada where his group was a pioneer in using bacterial genomics to study outbreaks, including the large Canadian listeriosis outbreak in 2008 and then the Haitian cholera outbreak of 2010. With this experience in public health, from 2015 to 2020 Matthew was the Scientific Director General of Canada’s National Microbiology Laboratory. At the NML, Matthew had significant leadership roles for pandemic preparedness and response, as he was also co-chair of the Canadian Public Health Laboratory Network and the Global Health Security Action Group Laboratory Network.
Matthew also has clinical laboratory expertise, as he previously served as a Clinical Microbiologist at the Health Sciences Centre in Winnipeg, where he was the laboratory lead for Infection Prevention & Control, and he remains interested in the evolution and transmissibility of antibiotic resistant organisms.
As a culmination of these experiences, Matthew is also now Director of the Food Safety Research Network, based at the Quadram Institute. This network has the goal of brokering collaborative research projects between food businesses and academic research groups that will make UK foods safer from microbial risks.
Professor David Graham is Professor of Ecosystems Engineering at Newcastle University who has led AMR surveillance and related projects around the world for over 30 years. He is a member of the United Nations (UN) Quadripartite Technical Group on Integrated Surveillance on Antimicrobial Use and Resistance and is the integration lead on the new Technical Guide on cross-sectoral AMR surveillance for member states. Graham was co-lead author on the UN report, “Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance” and co-wrote the World Health Organisation recommendations aimed at improving water quality, sanitation, and hygiene (WASH) as a strategy for reducing AMR in the low-resource settings. Domestically, Graham worked on UK SAGE sub-group on COVID-19 Transmission in the Wider Environment and was in the Expert Advisory Group who operationalized UK wastewater surveillance during the COVID-19 pandemic. He is a Chartered Engineer, but uses microbiology, genetics, and genomics to create holistic solutions. His current work focuses on relationships between human behaviour, water security, AMR, and sustainable development.
Dr Edward Haynes is the PATH-SAFE Science Fellow. He is a molecular biologist by training, with a background in bacterial genomics of foodborne, plant and honeybee pathogens. His current role is shared between his molecular biology responsibilities at Fera Science and PATH-SAFE Science Fellow. Earlier in his career, he held another joint post between Fera and the FSA to develop genomic approaches to foodborne bacteria, which involved working with the US Food and Drug Administration (FDA) to learn from the extensive work they had been doing to routinely apply genomic tools to foodborne pathogens in the GenomeTrakr project. In the last few years, he has also led projects applying non-targeted metagenomic sequencing approaches to detect AMR genes in different parts of the food production chain, in meat factories and in ready-to-eat foods.
Professor Rene S. Hendriksen is a professor at the Technical University of Denmark, National Food Institute, and leads the Research Group of Global Capacity Building. He is the director of the reference centres; World Health Organization (WHO) Collaborating Centre, Food and Agriculture Organization (FAO) Reference Center, and European Union Reference Laboratory for Antimicrobial Resistance (AMR) and Genomics, respectively. In his early days, the focus was on conventional and molecular microbiology. Since 2010, he has embraced the era of genomics with a strong focus improving global surveillance of antimicrobial resistance through capacity building, and implementing research. In addition, he provide scientific and technical assistance including the provision of external quality assessment trials to supranational organizations, national reference laboratories, health organizations and foundations, globally.
Professor Jay Hinton did his first degree in Microbiology, his PhD at the University of Warwick and his postdoc at the University of Oxford’s Weatherall Institute of Molecular Medicine. Jay became Head of Molecular Microbiology at the Institute of Food Research in Norwich in 1999, and relocated to Trinity College Dublin in 2009. Jay moved his research group to the University of Liverpool in 2012, where he is currently the Professor of Microbial Pathogenesis. In 2003, Jay’s group pioneered a transcriptomic approach that revealed a “snapshot” of Salmonella gene expression during the process of infection of mammalian cells. They discovered a key mechanism for silencing gene expression in bacteria in 2006 mediated by the H-NS protein. At Trinity College Dublin, his team employed RNA-seq-based approaches to understand the transcriptome of Salmonella Typhimurium which involved the identification of 280 non-coding sRNAs. At the University of Liverpool, Jay’s research group is using functional genomics to understand how new Salmonella pathovariants are causing endemic bloodstream infections across sub-Saharan Africa. This disease, iNTS, has killed around 500,000 people over the past decade. His team is now using a combination of molecular microbiology, genomics and functional transcriptomics to generate new insights. Jay lead the 10,000 Salmonella genomes project (https://github.com/apredeus/10k_genomes), which sequenced bloodstream isolates from about 50 countries dating from 1949 to 2017, with ~80% representing African and Latin-American datasets. Recent discoveries include the determination of the role of a single noncoding nucleotide in the over-expression of a key virulence factor in African Salmonella (Hammarlöf et al, PNAS) and an understanding of the step-wise evolution of African Salmonella (Pulford et al, Nature Microbiology).
Professor Kat Holt is a computational biologist specialising in infectious disease genomics. She is Professor of Microbial Systems Genomics at the London School of Hygiene and Tropical Medicine (LSHTM) Department of Infection Biology, and Co-Director of the LSHTM AMR Centre. She is also an Adjunct Professor (Research) at Monash University’s Department of Infectious Diseases in Melbourne, Australia; a HHMI-Gates International Research Scholar; and former Editor-in-Chief of the UK Microbiology Society journal Microbial Genomics. Kat has a BA/BSc from the University of Western Australia majoring in Biochemistry, Applied Statistics and Philosophy, with Honours in Genetics (2004); a PhD in Molecular Biology from the University of Cambridge and the Wellcome Trust Sanger Institute on the genomics of typhoid fever; and a Masters in Epidemiology from the University of Melbourne (2011). She has held Early Career (2010-2013) and Career Development (2014-2017) Fellowships from the NHMRC of Australia, and a Senior Medical Research Fellowship from the Viertel Foundation of Australia (2018-2021). Kat and her research team are particularly interested in the global health crisis of AMR, using genomic epidemiology tools to understand the evolutionary history and global dissemination of multidrug resistant pathogens, and developing new tools for prospective surveillance and tracking of emerging problems in the public health and clinical infectious disease space. Kat is a founder and coordinator of the Global Typhoid Genomics Consortium and the KlebNET Genomic Surveillance Platform.
Dr Claire Jenkins started working for UKHSA, formerly PHE, as a Clinical Scientist in 1996. She became head of the E. coli Reference Laboratory in 2012 and deputy head of the Gastrointestinal Bacteria Reference Unit in 2014, the same year as WGS was implemented as the routine typing method for surveillance and outbreak investigation.
Dr Keith Jolley is a bioinformatician working at the Department of Biology, University of Oxford, with Professor Martin Maiden. He has a PhD in Biochemistry from the University of Bath where he graduated in 1996. Following a PostDoc in Southampton, Keith moved to Oxford in 1998 to work on molecular typing of meningococcal carriage populations and gained programming and bioinformatics experience. Keith has been developing and overseeing the PubMLST website and the underlying molecular typing and genomic platforms for over 20 years (BIGSdb being the current iteration). He is also the co-developer of the ribosomal MLST scheme that can be used to identify and type all bacteria. The PubMLST website that he oversees hosts molecular typing schemes for more than 130 microbial species, serving as a nomenclature server for many third-party applications, and contains structured curated data for over 1.2 million bacterial genomes.
Professor Jason King is Advanced Vice-Chancellors Fellow and Royal Society University Research Fellow at the University of Sheffield.
Professor Robert A. Kingsley is a Professor of Microbiology at the University of East Anglia and a group leader and Principal Investigator at the Quadram Institute in Norwich, UK. His team’s work combines bioinformatics, genomics, molecular microbiology, and models of infection to investigate interactions between pathogens, the environment, and the host. Genomic diversity of bacterial pathogens and associated microbial and viral communities are studied to understand the function and evolutionary history of genes important to disease, transmission, drug resistance and phage sensitivity. Computational analysis generates hypotheses that are tested using classical microbiology, molecular genetics and models of infection. Our work has shed light on how new pathogens emerge and evolve, how epidemics spread and factors affecting risk to human and animal health. The long-term objective is to use this insight to improve policy decisions and in the rational design of intervention strategies, surveillance, and risk assessment of foodborne pathogens.
Dr Philippe Lehours has held a professorship at Bordeaux University Medical School and Bordeaux University Hospital since 2016. He is a pharmacist and holds a PhD in microbiology. During the period 2016-2021 he led an INSERM team focused on understanding the pathophysiological mechanisms of gastric carcinogenesis induced by Helicobacter pylori infection. Since 2017, he has headed the National Reference Center for Campylobacter and Helicobacter (NRCCH). At the NRCCH, he has routinely developed the sequencing of mainly Campylobacter genomes in order to study their phylogeny, resistome and virulome, and identify source attribution markers. NGS-type approaches directly from biological samples are also under development.
Dr Kirsty Line is the Head of the Surveillance and Laboratory Services Department in the Science Directorate of the Animal and Plant Health Agency (APHA) of Defra.
Professor Nick Loman works as Professor of Microbial Genomics and Bioinformatics in the Institute for Microbiology and Infection at the University of Birmingham. His research explores the use of cutting-edge genomics and metagenomics approaches to the diagnosis, treatment and surveillance of infectious disease. Nick has so far used high-throughput sequencing to investigate outbreaks of important Gram-negative multi-drug resistant pathogens, and recently helped establish real-time genomic surveillance of Ebola in Guinea. His current work focuses on the development of novel sequencing and bioinformatics methods to aid the interpretation of genome and metagenome scale data generated in clinical and public health microbiology.
Professor Martin Maiden is a molecular microbiologist. He attended the University of Reading (BSc, Microbiology with subsidiary Chemistry) as an undergraduate and the University of Cambridge as a Postgraduate (PhD, Biochemistry) and an MRC Training Fellow. He worked for nine years in the NHS (National Institute for Biological Standards and Control), including a one-year sabbatical in Germany (Max Planck Institut für Molekulare Genetic). He joined the University of Oxford in 1997 as a Wellcome Trust Senior Fellow. He has been a faculty member (Departments of Zoology and Biology), College Fellow (Hertford College), and Professor since 2004 and was Senior Proctor of the University of Oxford 2019-2020. From October 2024 he shall serve as Head of the Department of Biology, University of Oxford. For forty years he has applied molecular approaches to studying bacteria, with an emphasis on translation to infectious disease control, especially vaccination, with a strong emphasis on open access data. He is a committed educator and academic administrator.
Professor Alison Mather completed a BSc (biomedical toxicology) and MSc (epidemiology) at the University of Guelph in Canada, followed by a PhD in epidemiology at the University of Glasgow. She then worked at the Wellcome Sanger Institute in pathogen genomics before moving to the University of Cambridge for a BBSRC Fellowship on the genomic epidemiology of Salmonella and antimicrobial resistance (AMR) in animals and humans. In October 2017 she became a Group Leader at Quadram Institute Bioscience (QIB) and the University of East Anglia (UEA); currently, she is the co-Lead of the Microbes and Food Safety Institute Strategic Programme at QIB and an Honorary Professor in the Norwich Medical School at UEA. Her research interests are to understand the origins, evolution and transmission of foodborne bacteria, with a focus on bacteria that are resistant to antimicrobial drugs.
Dr Marie McIntyre is the NUPAcT Fellow in Translational Food Safety in partnership with the FSA, Marie’s human and veterinary public health research focuses on the drivers of and control measures for infectious disease, particularly using a One Health ethos. She led a systematic review examining the sensitivity to climate of high impact human and domestic animal pathogens, identifying food-, soil-, water- (and vector-)borne transmission routes as particularly associated with climate. She managed field work and data collation on the ‘Fully integrated, real-time detection, diagnosis and control of community diarrhoeal disease clusters and outbreaks’ (Integrate) project, and led development of literature reviewing processes for the Global Burden of Animal Diseases programme. Recently, she has been exploring ideas to collate evidence for antimicrobial resistance in the environment using big data approaches. Within her fellowship, Marie aims to provide policy-appropriate ‘living’ evidence collation and synthesis, and risk assessment tools and mechanisms to aid the FSA’s role in ensuring food for consumption is safe from foodborne disease and food supplies are sustainable.
Professor Frank Møller Aarestrup’s research has primarily targeted the association between use of antimicrobial agents to farm animals and the emergence and spread of antimicrobial resistance including the human health consequences.It has become increasingly clear that bacteria do not respect traditional borders and with the increased globalisation a problem in one country has become a problem for all countries. Thus, the research has increasingly been directed towards the global spread of initially foodborne, but now also other pathogenic bacteria. His research has contributed to the international standards for detection and monitoring of antimicrobial resistance in food borne pathogens and had major influence on the ways antimicrobial agents are used worldwide. The global focus is also documented by the fact that the research has been conducted with more than 400 co-authors, in more than 135 institutions in more than 35 countries. Furthermore, DTU-Food has been appointed WHO and EU reference laboratory for antimicrobial resistance in foodborne pathogens.
Dr Shateesh Nair is Senior Scientist at UKHSA working on WGS of gastrointestinal bacteria.
Professor Sascha Ott is an expert in developing targeted computational methods addressing biomedical needs. He is leading a research group at Warwick Medical School and a Bioinformatics service unit called Bioinformatics Research Technology Platform (RTP) with currently seven permanent staff members. His lab developed the “Wellington” footprinting methodology and software for high-throughput open chromatin data sets (Nucleic Acids Research 2015) which has found wide impact in cancer research, immunology, and a range of other biomedical settings, and a new method for the analysis of DNA methylation data which led to the hypothesis that stem cells may be depleted in the endometrium of recurrent miscarriage patients. His lab contributed to the design of the urine-based bladder cancer test GALEAS Bladder marketed by Informed Genomics. Since the retirement of Prof. Mark Achtman, Sascha has been responsible for the maintenance and development of the EnteroBase system by the Bioinformatics RTP team. He is PI on the BBSRC-funded PhytoBacExplorer project, developing an EnteroBase-like resource for the phytobacterial community. He also currently holds a joint Wellcome Trust Investigator award with Prof. Jan Brosens, and is co-investigator on one CMMI-EPSRC and one MRC-funded project.
Associate Professor Torsten Seemann is a world-renowned Bioinformatician who has developed cutting edge analysis approaches to enhance the use of genomic data for Discovery Science and Public Health. He is the Lead Bioinformatician at the Centre for Pathogen Genomics and the Director of Bioinformatics at Doherty Applied Microbial Genomics. His expertise includes the management of hardware and the design of software analysis infrastructure required to interrogate pathogen genome data to understand pathogen evolution, transmission and drug-resistance. He also led national and international bioinformatic analysis for SARS-CoV-2 during the COVID-19 pandemic as well as the development and deployment of Australia’s first real-time data-sharing and national genomics surveillance platform, AusTrakka. Torsten has been a significant contributor to international genomics standards through the Public Health Agency for Genomics Epidemiology (PHA4GE) and has recently been appointed as Lead Bioinformatics for the US CDC’s PulseNet International Asia-Pacific Initiative.
Dr Eric L. Stevens is an International Policy Analyst in the Office of the Center Director at the Center for Food Safety and Applied Nutrition. He received his Ph.D. in Human Genetics and Molecular Biology from The Johns Hopkins School of Medicine with an emphasis on human population genetics and estimating genetic relatedness. Dr. Stevens works closely with international organisations and our foreign regulatory counterparts on food science and safety issues. Additionally, Dr. Stevens is the Codex Alimentarius manager for CFSAN and helps coordinate FDA’s participation with the US Codex Office and at Codex Committee Meetings.
Dr Maria Traka is the deputy Head of Food & Nutrition National Bioscience Research Infrastructure (F&N-NBRI), based at Quadram Institute Bioscience (QIB). F&N-NBRI is a national coordinating ‘hub’ in food, nutrition and health and the leading national provider of new and continuously updated data, tools and services vital for public health, research and innovation. Maria’s research focus is understanding the role of plant-rich diets in improving health. She has an active interest in using data-driven approaches in personalized nutrition, prevention of non-communicable diseases, and the importance of the microbiome in modulating response to complex diets.
Maria actively participates in international efforts to coordinate FAIRdata in nutrition. She was a PI in the Food and Nutrition Security Cloud (FNS Cloud, Horizon2020, 2019-2022; https://www.fns-cloud.eu/) leading the technical delivery of linking diet and microbiota datasets available in public resources, which led to the development of a metadata search tool for diet and microbiome, Fairspace, as well as the development of the FNS-ontology. Maria is also leading work within the ELIXIR Food and Nutrition community on diet and microbiome data interoperability. She has been involved and led in nationally and internally funded projects around diet and health.
Maria is also the co-Director of the Food Safety Research Network, based at QIB, which connects food industry, food and health policymakers and academia through collaborative research to protect the UK from foodborne hazards.
Dr Adriana Vallejo-Trujillo is a researcher in WGS of microbial pathogens at the Roslin Institute, University of Edinburgh.
Dr Nicole Wheeler is a Research Fellow at the University of Birmingham. Her work focuses on the development of computational approaches to addressing biosecurity problems. She has provided expertise on machine learning for genomic pathogen and AMR surveillance for several international initiatives and is actively involved in the development of governance frameworks to ensure the safe and responsible development of technologies for health improvement.
Dr Koji Yahara is a Group Leader at the Antimicrobial Resistance Research Center, National Institute of Infectious Diseases (a WHO Collaborating Center for AMR surveillance and research) in Japan. He is a bio- and medical-informatician responsible for Japan’s national AMR surveillance systems. He earned his Ph.D. in Biostatistics through a collaborative program bridging biostatistics and infectious disease epidemiology. In 2012, he began a long-term research stay in Germany (at the Max Planck Institute) and the UK (collaborating with Swansea, Oxford, and Imperial College London) as a government-funded JSPS Research Fellow from the University of Tokyo, contributing to international research projects on bacterial population genomics. Upon joining the National Institute of Infectious Diseases in April 2016, he assumed responsibility for the information technology of the Japan Nosocomial Infections Surveillance (JANIS), a comprehensive national AMR surveillance program. Since 2019, his team has been developing genomic surveillance programs and studies, as reported in Genome Medicine (2021) and Nature Communications (2023). His primary focus lies in computational and statistical analyses of genomic and epidemiological data related to infectious diseases, as well as in the development and management of information technology programs and databases for the national surveillances. He also serves as an Editor for Microbial Genomics.



.png)
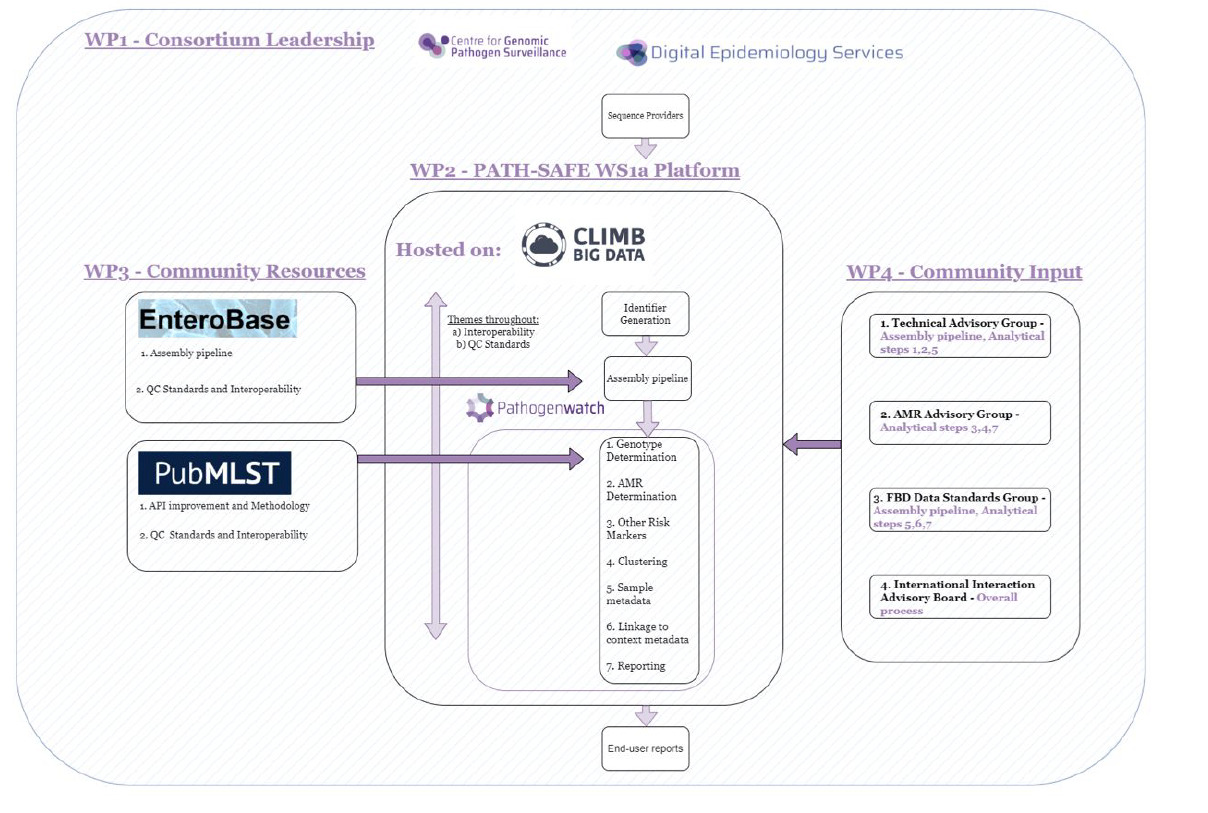

.png)